Icariin promotes expression of PGC-1α, PPARα, and NRF-1 during cardiomyocyte differentiation of murine embryonic stem cells in vitro1
Introduction
Mitochondrial biogenesis requires coordinated changes in the metabolic enzymes of oxidative phosphorylation, the TCA cycle, and fatty acid oxidation. The expression of hundreds of nuclear-encoded mitochondrial biogenesis-related genes is coregulated by a few nuclear transcription factors and co-activators. Nuclear respiratory factor (NRF)-1 and NRF-2 regulate many of the genes encoding oxidative phosphorylation proteins[1,2]. Peroxisome proliferator-activated receptors (PPAR) regulate genes encoding enzymes and transporters of fatty acid oxidation[3-6]. Peroxisome proliferator-activated receptor γ coactivator-1 alpha (PGC-1α) is a nuclear-encoded transcriptional co-activator which plays a critical role in the control of mitochondrial biogenesis based on its ability to interact with transcription factors that activate nuclear genes encoding mitochondrial proteins[7-9]. Accumulating evidence supports a major role for NRF-1 in mediating the effects of PGC-1α on mitochondrial biogenesis. PGC-1α can interact specifically with NRF-1 through the NRF-1 DNA binding domain. It can transactivate the transcription of NRF-1 target genes involved in mitochondrial respiration and induces NRF-1 mRNA[10]. In addition to its activation of respiratory subunit genes, PGC-1α can also upregulate genes involved in the mitochondrial fatty acid oxidation pathway through co-activating PPARα[11]. PGC-1α and PPARα are abundantly expressed in tissues with high oxidative energy demands, such as cardiac and skeletal muscles[12].
The p38 mitogen-activated protein kinase (MAPK) serves as upstream events to regulate PGC-1α both at the transcriptional and post-transcriptional level. For example, the activation of the p38 MAPK was involved in exercise which stimulates PGC-1α transcription in skeletal muscles[13]. The p38 MAPK can phosphorylate PGC-1α in 3 residues (T262, S265, and T298)[13] and leads to increased stability and half-life[14]. PGC-1α could also be regulated through the p38 MAPK-sensitive interaction with the repressor p160 Myb-binding protein[15].
Icariin (Figure 1) is an active ingredient of plant herb Epimedium, which possesses many kinds of biological actions, improving cardiovascular function, hormone regulation, immunological function modulation, and antitumor activity[16]. Using a model system comprised of embryonic stem (ES) cells, our previous work demonstrated that icariin significantly stimulated the cardiac differentiation of ES cells in vitro and resulted in increased and accelerated gene expression of α-myosin heavy chain (α-MHC) and myosin light chain 2 (MLC2v)[17,18]. Since the cardiomyocyte differentiation of ES cells in vitro faithfully replicates the process in vivo and ES cell-derived cardiomyocytes display properties similar to those observed in vivo or in primary cultures[19,20], the present study was designed to address the modulation of the most common factors (PGC-1α, PPARα, and NRF-1) implicated in the control of mitochondrial biogenesis by icariin during cardiomyocyte differentiation. In addition, the activation of the p38 MAPK was evaluated as it may be partly responsible for the effect of icariin.
Materials and methods
Cell culture and differentiation The permanent ES cell line D3 (CRL-1934, American Type Culture Collection, Manassas, VA, USA) was cultivated in an undifferentiated state on primary cultures of mouse embryonic fibroblasts in Dulbecco’s modified Eagle’s minimal essential medium (DMEM, Gibco BRL, Life Technologies, Germany), supplemented with 10% fetal calf serum (FCS, Gibco BRL, Germany), 0.1 mmol/L beta-mercaptoethanol (Sigma, St Louis, MO, USA), non-essential amino acids (NEAA, stock solution diluted at 1:100, Hyclone, Logan, UT, USA) and 106 units/L recombinant mouse leukemia inhibitory factor (Chemicon, Temecula, California, USA). For the differentiation of ES cells, embryoid bodies (EB) were generated using the hanging drop method with small modifications[21,22]. On d 0, 30 μL of drops containing approximately 600 ES cells were placed on the lids of Petri dishes filled with D-Hanks’ solution and cultivated in hanging drops for 3 d followed by another 2 d in the Petri dishes. On d 5, the EB were plated separately onto gelatin-coated, 24-well culture plates in differentiation medium that consisted of DMEM, 20% FCS, 0.1 mmol/L mercaptoethanol, and 1% NEAA. After incubation for 24 h (d 6), outgrown EB were subjected to 10-7, 10-8, and 10-9 mol/L icariin, respectively. 10-8 mol/L retinoic acid (RA) was used as the positive control.
Reagents Icariin was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (China; Batch N
Immunofluorescence analysis Differentiated EB that had been grown on coverslips were fixed for 20 min in methanol at -20°C, followed by permeabilization in 0.1% Tween 20 in phosphate-buffered solution (PBS). After washing in PBS 3 times, the EB on coverslips were transferred to PBS containing 10% goat serum (Sigma, USA) for 30 min at room temperature. The EB were then placed into PBS containing mouse monoclonal anti-α-actinin (Sigma, USA; dilution 1:100) and incubated overnight at 4 °C. The EB were washed in PBS 3 times, followed by incubation in PBS containing the FITC-conjugated antimouse IgG (Sigma, USA; dilution 1:500). Fluorescence recordings were performed by means of confocal laser scanning setup (Leica TCS SP2, Bensheim, Germany) connected to an inverted microscope.
Semiquantitative RT-PCR Total RNA was isolated from ES cells and the EB using Trizol reagent (Gibco BRL, Germany) in accordance with the manufacturer’s instructions. To synthesize first-strand cDNA, 1 µg total RNA was incubated with 0.5 µg of oligo (dT) 6 primer (Sangon, Shanghai, China) and 5 µL deionized water at 65 °C for 15 min. Reverse transcription reactions of 20 µL were performed with 200 units of M-MuLV reverse transcriptase (Gibco BRL), 4 µL of 5×reaction buffer, and 1 mmol/L deoxynucleoside triphosphate (dNTP) mixture for 1 h at 42 °C. PCR of 50 µL contained 1 µL of the RT reaction product, 5 µL of 10×PCR buffer, 25 units Taq polymerase (Sangon, China), 1 µL of 10 mmol/L dNTP mixture, and 30 pmol of each primer.
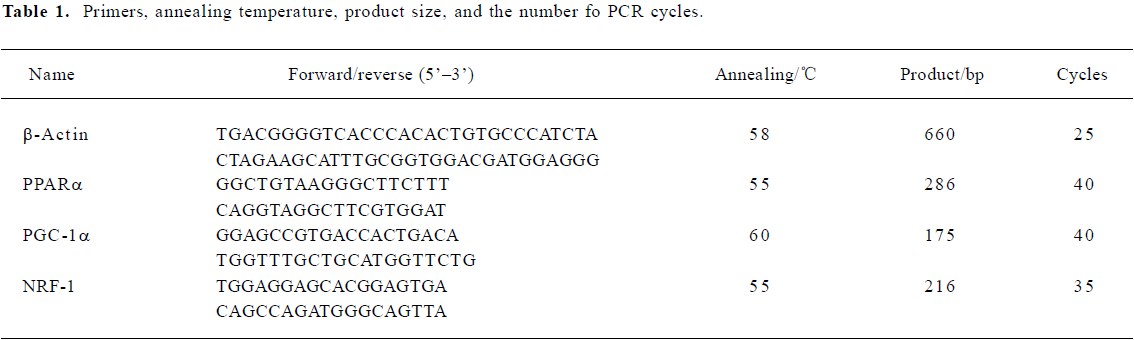
Primers, annealing temperature, product size, and the number of PCR cycles are depicted in Table 1. The PCR products were analyzed by 1.5% agarose gel electrophoresis, visualized with ethidium bromide staining, and then quantified using a bio-imaging analyzer (Bio-Rad, USA). The density of the products was quantified using Quantity One version 4.2.2 software (Bio-Rad, USA). β-Actin was used as an internal standard.
Full table
Western blot analysis The cells were washed with PBS, collected in RIPA buffer (containing 0.2% Triton X-100, 5 mmol/L EDTA, 1 mmol/L PMSF, 10 µg/mL leupeptin, and 10 µg/mL aprotinin) and lysed for 30 min on ice. The aliquots were assayed for protein concentration using the Bio-Rad protein assay kit and equal amounts of protein were loaded per well on a 12% SDS–PAGE. Subsequently, the proteins were transferred onto 0.45 µm pore size nitrocellulose membranes and blocked with blotto (5% dry milk in PBS, pH 7.4, with 0.1% Tween 20) at room temperature.
The blots were challenged with primary antibody in blotto overnight at 4 °C, followed by washing 3 times with PBST (0.1% Tween 20) at room temperature and challenged with horseradish peroxidase-conjugated goat anti-rabbit, rabbit anti-goat, or mouse anti-mouse antibodies (Affinity Bioreagents, Golden, CO, USA; dilution 1:1000), respec-tively, followed by detection with an enhanced chemiluminescent substrate (Pierce, USA). As primary antibodies, the goat polyclonal anti-actin, the mouse monoclonal anti-troponin T, rabbit polyclonal anti-PGC-1α, rabbit polyclonal anti-PPARα, rabbit polyclonal anti-NRF-1 (Santa Cruz Biotechno-logy, Santa Cruz, CA, USA; dilution 1:500), and the mouse monoclonal anti-α-actinin (Sigma–Aldrich; dilution 1:500) the rabbit polyclonal anti-p38 MAPK, and anti-p-p38 MAPK (Cell Signaling, USA; dilution 1:1000) were used.
Statistics Student’s t-test and one-way ANOVA were used to determine the statistical significance of differences between values for various experimental and control groups. P<0.05 was considered statistically significant.
Results
In vitro cardiomyocyte differentiation of ES cells The attached culture was established by plating a single, d 5 EB culture onto a 24-well plate and allowing continued cellular proliferation and differentiation. Within this multicellular arrangement in the EB outgrowths, cardiomyocytes appeared as spontaneously contracting, round cell clusters within the EB on average 3 d later (d 8). An increase in size, strength of contraction, and beat frequency was observed during further differentiation. Cardiomyocytes derived from ES cells were positive for the α-actinin antibody, and cross striations were observed at higher magnification (Figure 2A). A mass increase in expression of α-actinin and troponin T was detected on d 10 (Figure 2B).
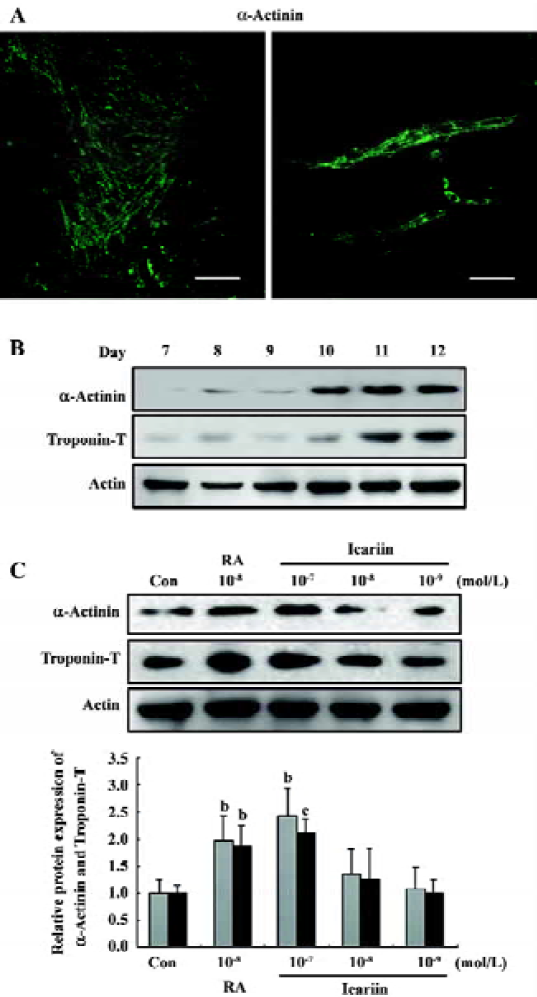
After incubation for 24 h (d 6), the attached EB were subjected to 1×10-7, 1×10-8, and 1×10-9 mol/L icariin, respectively, and cardiac sarcomeric proteins were evaluated on d 12. It was apparent that the protein level of α-actinin and troponin T in the EB was dose-dependently upregulated by icariin exposure (Figure 2C).
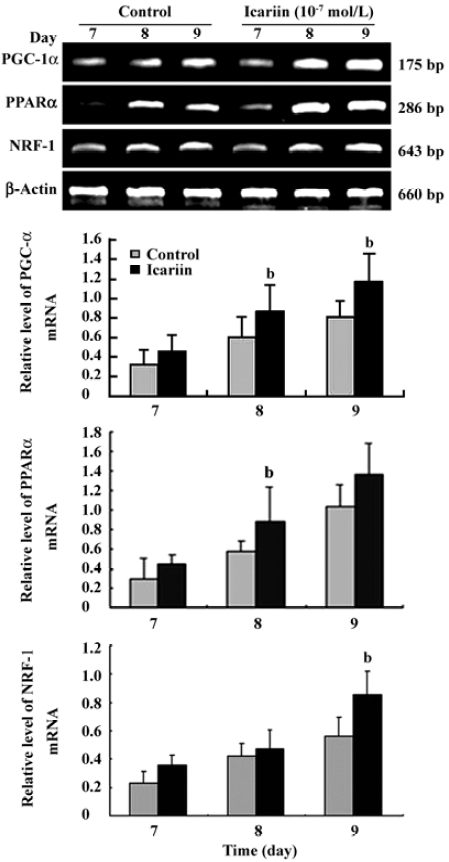
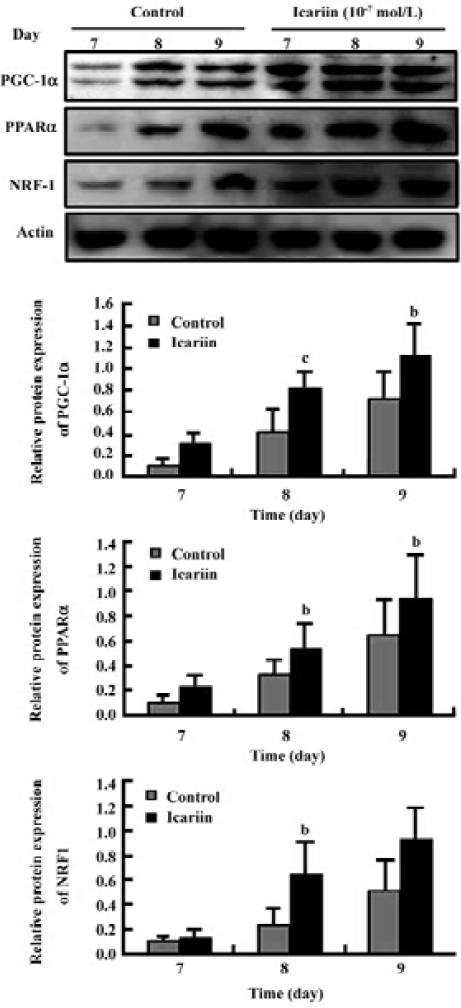
Transcription analysis of PGC-1α, PPARα, and NRF-1 during cardiomyocyte differentiation Semiquantitative RT-PCR was employed to elucidate the pattern of PGC-1α, PPARα, and NRF-1 gene expression during the differentiation course. The data showed that PGC-1α, PPARα, and NRF-1 mRNA levels increased obviously in early differentiation and the prominent changes took place between d 7 and d 9. Compared to the control group, the mRNA levels of PGC-1α, PPARα, and NRF-1 were markedly upregulated when icariin was present (Figure 3).
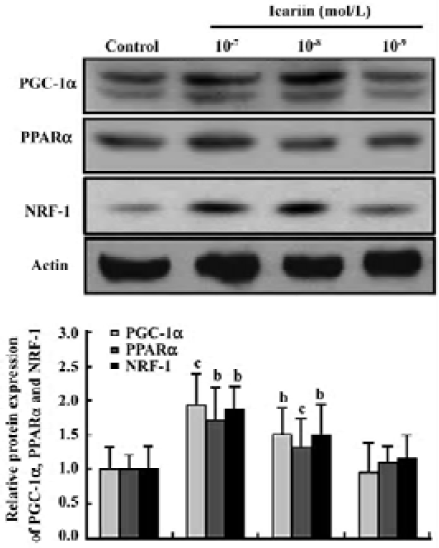
Protein analysis of PGC-1α, PPARα, and NRF-1 during cardiomyocyte differentiation To confirm the result of the gene expression found by semiquantitative RT-PCR, the overall level of protein expression in the EB during cardio-myocyte differentiation was analyzed. Different concentrations of icariin were applied to confirm the effect of icariin on the expression of PGC-1α, PPARα, and NRF-1. Showing an analogy with mRNA expression, the protein level of PGC-1α, PPARα, and NRF-1 increased in early differentiation (Figure 4). This prominent expression abated before the mass expression of α-actinin and troponin T, suggesting that PGC-1α, PPARα, and NRF-1 may be critical to normal cardiac develop-ment in vitro. Moreover, elevations in the protein expression of PGC-1α, PPARα, and NRF-1 were enhanced by icariin in a dose-dependent-manner (Figures 4, 5).
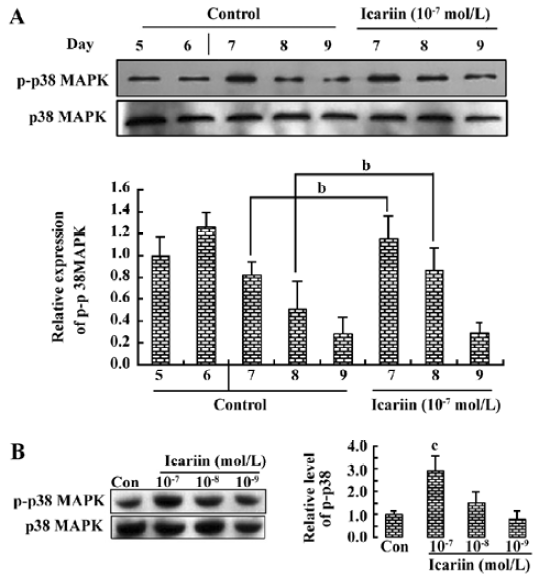
Involvement of p38 MAPK activation in icariin-induced cardiomyocyte differentiation We hypothesized that icariin relayed p38 MAPK activity to the transcription of genes involved in cardiomyocyte commitment. The phosphorylation of the p38 MAPK was further activated and prolonged by icariin in early differentiation. In the absence of icariin treatment (control), p38 MAPK activity peaked spontaneously on d 6 and decreased on d 8, while in the presence of icariin, the p-p38 MAPK was maintained at a high level until d 8 (Figure 6A). Moreover, the activation of the p38 MAPK by icariin was in a dose-dependent manner (Figure 6B).
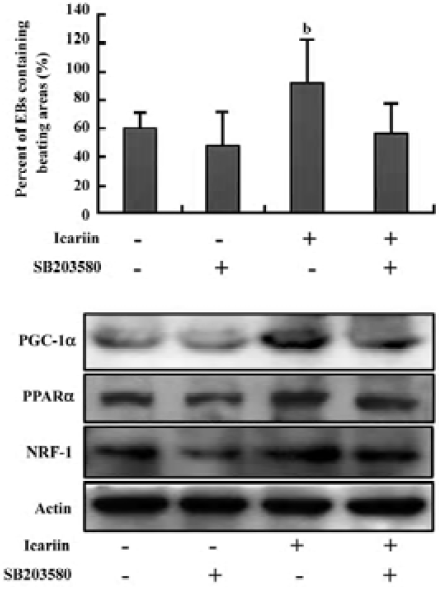
To further investigate the impact of p38 MAPK activity on icariin-enhanced cardiomyocyte differentiation and the expression of PGC-1α, PPARα, and NRF-1, the EB were treated with icariin alone or together with SB203580 (10 µmol/L) from d 6. Subsequently, cardiomyocyte differentiation was assessed by calculating the percentage of spontaneously-contracting EB on d 12. It was shown that icariin-stimulated cardiac differentiation was abolished by SB203580, while SB203580 alone had a slight effect on differentiation (Figure 7A). Similarly, the increase in the expression of PGC-1α, PPARα, and NRF-1 following treatment of icariin was inhibited in the presence of SB203580 (Figure 7B). These results implied that high p38 MAPK activity was associated with cardiogenesis and was responsible for icariin-induced cardio-myocyte differentiation.
Discussion
Cardiomyocyte differentiation can be divided in 2 processes: cardiogenesis and cardiac myofibrillogenesis. Early cardiogenesis is regulated by 3 families of transcription factors (ie Nkx2.5, MEF2C, GATA4), which start to be fully expressed on d 5. While cardiac transcription factors (ie α-MHC and MLC2v) encoding contracting proteins have not appeared at d 5. Thus, d 5 is a critical window at which the cardiac differentiation program becomes fully activated, but no cardiac cells identified by organized cardiac sarcomeric proteins or by functional automatic contractions are yet present[23]. Our previous study showed that there would be increasing and accelerating gene expression of α-MHC and MLC2v in EB when treated with icariin from d 5. In present study, the inducible effect of icariin was further demonstrated by evaluating cardiac sarcomeric proteins. The expression of α-actinin and troponin T were upregulated when the EB were subjected to icariin 24 h after transferring to 24-well plates, implying that icariin plays a role in myofibril-logenesis. It has been reported that treatment with high concentrations of RA (10-7 and 10-8 mol/L) between d 5 and d 7 would accelerate cardiomyocyte differentiation[24]. Therefore, RA was employed in this study as the positive control and its inducible effect was replicated as we expected.
Most studies on the control of mitochondrial gene expression implicate PGC-1α as a “master controller” of mitochondrial biogenesis, co-activating PPARα and NRF-1[7]. The conditions that provoke mitochondrial biogenesis, such as contractile activity, could induce the expression of PGC-1α[25]. Previous studies have demonstrated that the expression of the PGC-1α gene is greatly increased in the developing mouse heart, immediately before the large burst of mitochondrial biogenesis and the oxidative metabolism that precedes birth. Cardiomyocytes derived from murine ES cells display properties similar to those observed in cardiomyocyte in vivo or in primary cultures, expressing cardiac gene products in a developmentally-controlled manner, showing characteristic sarcomeric structures and possessing membrane-bound ion channels. Mitochondrial number and functional capacity in cardiomyocytes are dynamically regulated in accordance with energy demands during developmental stages and in response to diverse physiological conditions[26]. Here we showed that the protein expression of PGC-1α was induced in early differentiation, implying that the cardiomyocyte differentiation of ES cells was accompanied with an organization of mitochondrial biogenesis.
The expression of PGC-1α, PPARα, and NRF-1 was coincidently induced in early cardiomyocyte differentiation, representing a highly favorable environment for the onset of mitochondrial biogenesis in the EB. The parallel increase in NRF-1 ensured the coordinate induction of mtDNA transcription and replication, subsequently leading to the enhanced expression of mitochondrial proteins that are vital for respiratory chain function. In the transition from fetal to neonatal and adult life, cardiac metabolism switches from glucose to fatty acids as a preferred energy substrate to generate ATP[27]. This transition is accompanied by changes in activity and expression levels of several enzymes and regulators involved in fatty acid metabolism. The main regulators of fatty acid enzymes on the transcriptional level are the so-called PPAR which are up-regulated at birth[28,29]. In the present study, the expression of PPARα was observed to increase in early differentiation, which correlated to the increase in the expression of cardiac-specific transcription factors and proteins, indicating that in addition to the activation of suites of genes encoding contractile proteins, myofibrillogenesis is accompanied by an organization of enzymes involved in fatty acid metabolism. Interestingly, this phenomenon was recently replicated as the mRNA levels of NRF-1 and PPARα increased significantly during myogenesis in vitro[30]. It should be noted that PPARα relied upon ligands for activation. EB were grown in the presence of 20% fetal bovine serum during differentiation, so there would have been abundant natural ligands (eg fatty acids) for the activation of PPARα.
Mitochondrial biogenesis and the activation of both oxidative phosphorylation as well as the transcription and replication of the mitochondrial genome are key regulatory events in cell differentiation. The aim of this study is to analyze the mediation of mitochondrial biogenesis-related transcription factors by icariin, as final ES cell commitment may be influenced by mitochondrial proliferation and mtDNA transcription. The expression of PGC-1α, PPARα, and NRF-1 were upregulated by icariin in a dose-dependent manner during cardiac differentiation in vitro, suggesting that icariin facilitated mitochondrial adaptation to increase energy demand. Mitochondrial has proven to be important in maintaining the proper function of cardiomyocytes. During development of the heart, mitochondrial enzyme activities and proteins increase in the bovine and human heart[31]. The blockade of mitochondrial activity by the inhibition of mitochondrial protein synthesis, the uncoupling of the inner membrane potential from ATP synthesis and the inhibition of mitochondrial ATP production (oligomycin) inhibits differentiation of skeletal muscle cells from myoblast precursors[32].
The p38 MAPK was proposed to be a link between cardiogenesis and mitochondrial biogenesis. P38 MAPK activity stimulates PGC-1α gene transcription in cardiomyocytes, and the activation of this pathway is sufficient to induce, and is necessary for, cardiac muscle adaptation. In addition, the p38 MAPK could directly phosphorylate PPARα and then drive its own transcription in a ligand-dependent manner[33]. Interestingly, accumulating evidence indicates that the p38 MAPK plays a critical role in cardiomyocyte differentiation of murine carcinoma stem cells and ES cells in vitro[34,35]. The control of p38 MAPK activity constitutes an early switch, committing ES cells into either neurogenesis (p38 off) or cardiomyogenesis (p38 on)[36]. Consistent with previous studies, the p-p38 MAPK was found to be at a high level from d 5 and followed with a decrease on d 8. Because PGC-1α is a direct downstream target of the p38 MAPK, the resulting phosphorylation and protein stabilization could be important in mediating the increase in the PGC-1α protein observed here. In this study, we demonstrated that the phosphorylation of the p38 MAPK was enhanced and prolonged by icariin, suggesting that icariin triggered the activation of the p38 MAPK and thus mediated cardiac differentiation.
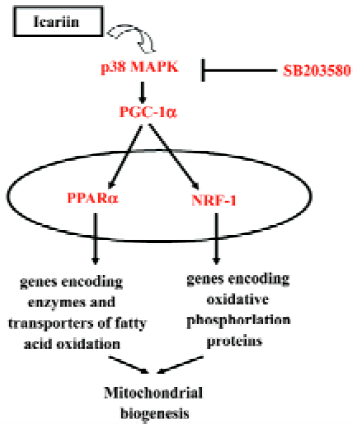
Taken together, icariin treatment stimulated the phosphorylation of the p38 MAPK and upregulated the expression of PGC-1α, PPARα, and NRF-1 (Figure 8) which may act as part mechanisms for the inducible effect of icariin on the cardiac differentiation of murine ES cells.
References
- Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 2002;286:81-9.
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta 2002;1576:1-14.
- Barger PM, Kelly DP. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc Med 2000;10:238-45.
- Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol Chem 1998;273:23786-92.
- Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA 1994;91:11012-6.
- Yu GS, Lu YC, Gulick T. Co-regulation of tissue-specific alternative human carnitine palmitoyltransferase Ibeta gene promoters by fatty acid enzyme substrate. J Biol Chem 1998;273:32901-9.
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 2003;24:78-90.
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab 2001;12:360-5.
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest 2000;106:847-56.
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999;98:115-24.
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 2000;20:1868-76.
- Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA 1994;91:7355-9.
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem 2005;280:19587-93.
- Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell 2001;8:971-82.
- Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci USA 2001;98:9713-8.
- He W, Sun H, Yang B, Zhang D, Kabelitz D. Immunoregulatory effects of the herba Epimediia glycoside icariin. Arzneimittel-forschung 1995;45:910-3.
- Zhu D, Qu L, Zhang X, Lou Y. Icariin-mediated modulation of cell cycle and p53 during cardiomyocyte differentiation in embryonic stem cells. Eur J Pharmacol 2005;514:99-110.
- Zhu DY, Lou YJ. Inducible effects of icariin, icaritin, and desmethylicaritin on directional differentiation of embryonic stem cells into cardiomyocytes in vitro. Acta Pharmacol Sin 2005;26:477-85.
- Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res 2002;91:189-201.
- Miller-Hance WC, LaCorbiere M, Fuller SJ, Evans SM, Lyons G, Schmidt C, et al. In vitro chamber specification during embryonic stem cell cardiogenesis. Expression of the ventricular myosin light chain-2 gene is independent of heart tube formation. J Biol Chem 1993;268:25244-52.
- Wobus AM, Wallukat G, Hescheler J. Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers. Differentiation 1991;48:173-82.
- Metzger JM, Lin WI, Samuelson LC. Vital staining of cardiac myocytes during embryonic stem cell cardiogenesis in vitro. Circ Res 1996;78:547-52.
- Wei H, Juhasz O, Li J, Tarasova YS, Boheler KR. Embryonic stem cells and cardiomyocyte differentiation: phenotypic and molecular analyses. J Cell Mol Med 2005;9:804-17.
- Wobus AM, Kaomei G, Shan J, Wellner MC, Rohwedel J, Ji G, et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardio-myocytes. J Mol Cell Cardiol 1997;29:1525-39.
- Irrcher I, Adhihetty PJ, Sheehan T, Joseph AM, Hood DA. PPARgamma coactivator-1alpha expression during thyroid hormone- and contractile activity-induced mitochondrial adapta-tions. Am J Physiol Cell Physiol 2003;284:C1669-77.
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol 1988;4:289-333.
- Makinde AO, Kantor PF, Lopaschuk GD. Maturation of fatty acid and carbohydrate metabolism in the newborn heart. Mol Cell Biochem 1998;188:49-56.
- Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, -beta, and -gamma during rat embryonic development. Endocrinology 1998;139:2748-54.
- Steinmetz M, Quentin T, Poppe A, Paul T, Jux C. Changes in expression levels of genes involved in fatty acid metabolism: upregulation of all three members of the PPAR family (alpha, gamma, delta) and the newly described adiponectin receptor 2, but not adiponectin receptor 1 during neonatal cardiac development of the rat. Basic Res Cardiol 2005;100:263-9.
- Kraft CS, LeMoine CM, Lyons CN, Michaud D, Mueller CR, Moyes CD. Control of mitochondrial biogenesis during myogenesis. Am J Physiol Cell Physiol 2006;290:C1119-27.
- Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ. Heart mitochondrial DNA and enzyme changes during early human development. Mol Cell Biochem 2000;210:47-52.
- Herzberg NH, Middelkoop E, Adorf M, Dekker HL, Van Galen MJ, Van den Berg M, et al. Mitochondria in cultured human muscle cells depleted of mitochondrial DNA. Eur J Cell Biol 1993;61:400-8.
- Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem 2001;276:44495-501.
- Davidson SM, Morange M. Hsp25 and the p38 MAPK pathway are involved in differentiation of cardiomyocytes. Dev Biol 2000;218:146-60.
- Eriksson M, Leppa S. Mitogen-activated protein kinases and activator protein 1 are required for proliferation and cardio-myocyte differentiation of P19 embryonal carcinoma cells. J Biol Chem 2002;277:15992-6001.
- Aouadi M, Bost F, Caron L, Laurent K, Le Marchand Brustel Y, Binetruy B. p38 mitogen-activated protein kinase activity commits embryonic stem cells to either neurogenesis or cardio-myogenesis. Stem Cells 2006;24:1399-406.