A novel high-throughput format assay for HIV-1 integrase strand transfer reaction using magnetic beads1
Introduction
HIV-1 integrase (IN) is an important enzyme in the virus replication cycle and an attractive target for antiviral drug design because it catalyzes the integration of the virus genome into the human genome. IN catalyzes a 2-step reaction. The first step is the cleavage of 3' GT dinucleotide from each 3' end of the virus long terminal repeats (LTR) DNA (donor DNA). This reaction is termed 3' processing (3P) and depends on the recognition of the specific sequence at the U5 and U3 ends of LTR. In the subsequent step, strand transfer (ST), the 3'-processed DNA ends acting as nucleophilic agents, attack phosphodiester bonds in the host cellular DNA (target DNA) and splice these 3' ends into the target DNA. This reaction does not absolutely require any specific sequence within the host DNA[1–3].
During the past decade, in vitro assays for IN activities have been developed and applied for the screening of inhibitors. These assays include 2 categories in general: (i) low-throughput gel-based assays involving radioactively-labeled oligonucleotides[4,5]; and (ii) high-throughput microtiter assays using biotin (BIO) or digoxin (DIG)-labeled oligonucleotides with enzyme-coupled detection strategies for the quantification of the product[6–8]. Recently, a gel-based assay using microarrayed compound libraries and 1 microtiter assay based on time-resolved fluorescent resonance energy transfer have been developed[9,10]. These assays are established at the aim of high-throughput screening (HTS), high sensitivity, specificity, and reliability. Among these assays, solid-phase microtiter assays in which DNA were immobilized on the microplate surface were most widely used in the screening of antiviral compounds because they are fast, convenient, and are optimized in HTS format[6–8]. However, the need of plate coating and blocking in these assays is time and labor consuming, and the invalidation of plate blocking causes non-specific binding of DNA or antibodies on the microplate and subsequent higher background readings. Moreover, the immobilization of the DNA substrate on the microplate limits the application of these assays in reaction characters studies.
It is reported that small molecules that inhibit the 3P activity of IN in vitro show little antiviral activity in vivo, and the inhibition of ST activity is the primary key to antiviral efficiency in vivo[11,12]. IN is the only 1 of the 3 enzymes for which clinically useful drugs are not available to date. Therefore, it is important to develop in vitro assays for the detection of IN activities, especially ST activity, and apply these assays in the screening of antiviral compounds. In the present study, we describe a rapid and highly robust approach to HIV-1 IN ST activity using HTS technology. This assay was performed in a 96-well microplate. Based on the commercially-available magnetic beads, this assay is rapid, flexible, and specific, and there is no need for plate coating or blocking. Furthermore, 2 kinds of detection strategies, absorbance and fluorescence, have been conducted and 2 antiviral compounds were employed to test the effectiveness of this assay.
Materials and methods
Materials Oligonucleotide sequences corresponding to the U5 terminus of the HIV-1 LTR and the target substrate were synthesized and modified by Shanghai Sangon Bio (Shanghai, China): oligo I BIO-5' ACCCTTTTAGTCA-GTGTGGAAAATCTCTAGCA3', oligo II 5' ACTGCTAGA-GATTTTCCACACTGACTAAAAG 3', oligo III 5' ATGTGG-AAAATCTCTAGCGAT3'-DIG, and oligo IV 5' ATCGCTA-GAGATTTTCCACAT3'. Taq DNA polymerase was purchased from Dalian Takara Bio (Dalian, China); restriction endonucleases and T4 DNA ligase were from New England Biolabs (Beverly, MA, USA). Dynabeads M-280 streptavidin (SA) magnetic particle and the Dynal MPC-96B concentrator were purchased from Dynal Biotech (ASA, Oslo, Norway). The monoclonal anti-DIG alkaline phosphatase (AP) conjugate antibody and monoclonal anti-DIG fluorescein isothiocyanate (FITC) conjugate antibody were purchased from Sigma (St Louis, MO, USA). The nickel-chelating column (Chelating Sepharose Fast Flow) was purchased from Amersham Pharmacia Biotech (Piscataway, NJ, USA). HEPES, imidazole, β-mercaptoethanol, isopropyl-beta-D-thiogalactopyranoside, bovine serum albumin (BSA) and piperazine-N, N'-bis-2-ethanesulfonic acid (PIPES) were obtained from Merck (Hohenbrunn, Germany). Baicalein was purchased from Fluka (Buchs SG, Switzerland). All other chemicals were from Amresco (Solon, OH, USA).
Protein expression and purification The plasmid containing the HIV NL4-3 IN gene was used as the PCR template DNA. Site-directed mutagenesis was done by overlapping PCR to bring the F185K/C280S double mutations for the purpose of enhancing the protein solubility. The IN gene was modified to contain an Nde I site at the 5' end and a BamH I site and a termination codon (TAG) at the 3' end. The PCR product was digested with Nde I and BamH I and ligated with Nde I–BamH I-digested pET-28a to construct a pNL-IN recombinant plasmid. After confirmation by sequencing (Shanghai Sangon Bio, China), correctly constructed pNL-IN was expressed in Escherichia coli strain BL21 (DE3) as a soluble N-terminal 6-histidine tag fusion protein and purified by a nickel affinity column as described[13]. After elution from the nickel column, the purified protein was dialyzed against 1 mol/L NaCl, 20 mmol/L HEPES, pH 7.5, 0.1 mmol/L EDTA, 1 mmol/L dithiothreitol, and 10% (w/v) glycerol at 4 °C overnight and stored at -80 °C. SDS-PAGE was employed to analyze the expression and purification of the IN protein, and the IN concentration was measured by the Bradford method (Bio-Rad, Hercules, CA, USA).
Microplate ST assay For high-throughput purpose, ST assays were performed in a 96-well microplate (Corning, New York, NY, USA) in a final volume of 50 µL. The wells were washed once with 1×reaction buffer (25 mmol/L PIPES, pH 7.0, 10 mmol/L β-mercaptoethanol, 5% (w/v) glycerol, 0.1 g/L BSA, and 10 mmol/L MnCl2). In total, 800 ng IN was added and pre-incubated in reaction buffer. Subsequently, 1.5 pmol donor DNA and 15 pmol target DNA were added and the reaction was initiated. After incubation for 1 h at 37 °C, 1.5 mL magnetic particles (6.7×108 beads/mL) and 51.5 µL binding buffer (10 mmol/L Tris-HCl, pH 7.6, 2 mol/L NaCl, 20 mmol/L EDTA, and 0.1% [w/v] Tween 20) were added and incubated at 20 °C for 15 min. Then the wells holding the mixture were placed in a magnetic concentrator, the supernatant was discarded, and the wells were washed 3 times with phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST). Subsequently, 100 µL of 1:5000 diluted AP conjugate anti-DIG antibody was added and incubated for 30 min at 37 °C. Finally, the wells were washed 3 times with PBST and the magnetic beads were transferred into fresh wells; 100 µL P-Nitrophenyl Phosphate (P-NPP) substrate (0.1 mol/L Na2CO3, pH 9.5, 6.7 mmol/L P-NPP, and 2 mmol/L MgCl2) was added. The plates were read at 405 nm with a Model 680 microplate reader (Bio-Rad, USA).
Results
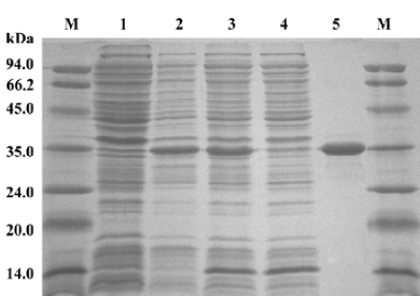
HIV-1 IN purification and characterization IN was purified from the soluble supernatant by nickel-affinity chromatography. The SDS-PAGE analysis indicated that the expression of recombinant IN exceeded the expression of any other cellular protein and IN was highly soluble. The molecular weight of the recombinant IN was approximately 33 kDa, with a purity of approximately 95% (Figure 1).
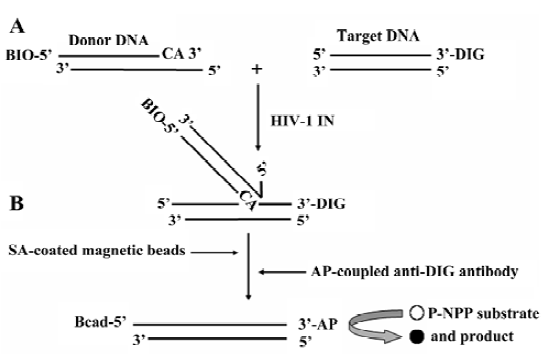
Principle of the HTS assay for HIV-1 IN ST reaction In this HTS assay, the 31 bp duplex donor DNA substrate was designed to mimic the U5 end of the HIV-1 LTR. After annealing, there was a 3 nucleotides overhang at the 5' end of oligo I for better interaction of BIO-SA in the following DNA capture step, and the 3' GT dinucleotides were removed from oligo I. The donor DNA was labeled at the 5' end of oligo I with BIO, and the target DNA was labeled at the 3' end of oligo III with DIG. The ST reaction resulted in a 5' BIO and 3' DIG-labeled covalent DNA product. The DNA product was captured by SA-coated magnetic beads through the specific BIO-SA interaction, followed by the addition of the AP conjugate anti-DIG antibody. Therefore, the product was measured by the AP-coupled enzyme reaction (Figure 2).
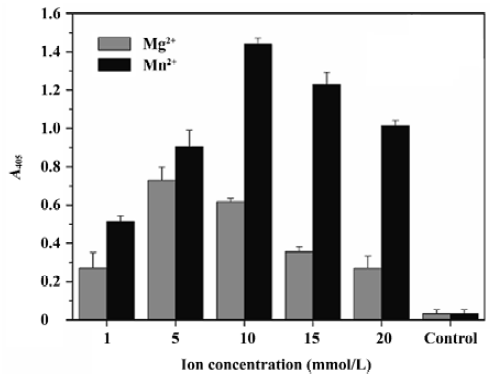
Optimization of the ST activity assay To optimize this HTS assay, we varied the concentrations of donor DNA, target DNA, metal ion, and the IN protein to obtain the optimal reaction conditions. The optimal concentration of donor DNA was 30 nmol/L. The IN concentration varied between 17 and 1000 nmol/L, and 300 nmol/L was determined to be optimal. As for the target DNA, we varied the concentration from 10 to 750 nmol/L, and determined that the 300 nmol/L target DNA to be added was optimal. Furthermore, we investigated the performance of our assay in the presence of either cationic cofactor, Mg2+ and Mn2+. Using this assay, ST activity peaked at 10 mmol/L Mn2+ and 5 mmol/L Mg2+, respectively. Accordingly, the presence of Mn2+ instead of Mg2+ increased IN activity by almost double (Figure 3). Here, 300 nmol/L IN, 30 nmol/L donor DNA, 300 nmol/L target DNA, and 10 mmol/L Mn2+ were determined to be the optimal reaction conditions and used throughout the subsequent study.
To test the sensitivity and specificity of this assay, we measured the signal of all reactions under optimal reaction conditions. Furthermore, 2 widely-used essential parameters to indicate the quality of an assay: the signal-to-noise ratio (S/N) and signal-to-background ratio (S/B) were defined and calculated as:
and
As a result, the negative controls in the absence of either IN, donor DNA, or target DNA showed background readings measured as absorbance at 405 nm (A405), lower than 0.05, whereas the ST reaction gave a signal of more than 1.4 (Figure 4A). Accordingly, the S/N and S/B ratios of the assay were 86 and 30, respectively.
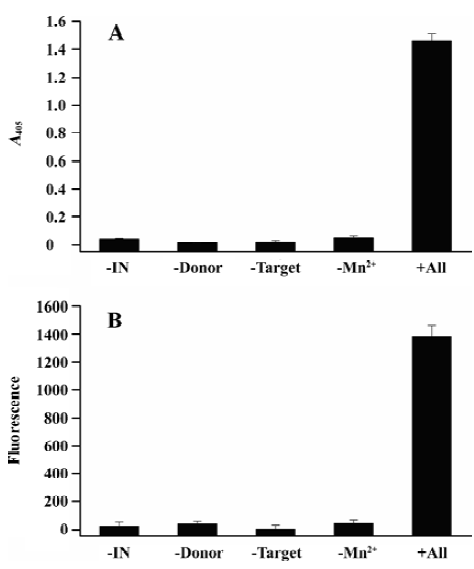
To further demonstrate the robustness of this assay, we changed the AP conjugate anti-DIG antibody to the FITC conjugate anti-DIG antibody. After capturing the reaction product by magnetic beads, 100 mL of 1:500 PBS-diluted FITC conjugate antibody was added and incubated for 30 min at 37 °C. Finally, the microplate was washed as described and the plates were read with a 1420 Multilabel counter VICTOR reader (Perkin Elmer, Boston, MA, USA) at 485 nm excitation and 535 nm emission. The results were similar to the assay in which the AP conjugate antibody was used. The ST reaction showed a fluorescence signal as high as almost 1400, whereas the signals of negative controls were lower than 40 (Figure 4B). The S/N and S/B ratios were 61 and 32, respectively.
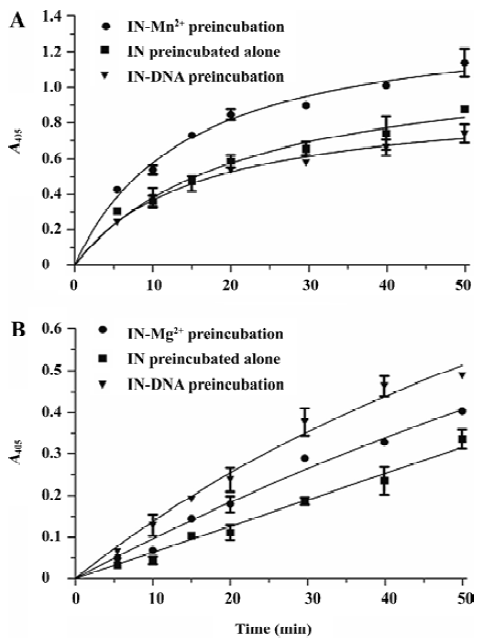
HIV-1 IN ST reaction character study The HIV-1 IN 3P reaction character was studied using the gel-based assay involving the radioactively-labeled oligonucleotide. Since IN pre-incubated in the presence of the DNA substrate was more active in the 3P reaction than IN pre-incubated alone or with Mg2+ [14], we were interested in the ST reaction character of IN under different pre-incubation conditions using our assay. We set 3 reaction groups: (i) IN pre-incubated alone in the reaction buffer without both DNA and Mn2+; (ii) IN pre-incubated with Mn2+ in the reaction buffer without DNA; and (iii) IN pre-incubated with DNA in the reaction buffer, but without Mn2+. The pre-incubation lasted for 30 min at 37 °C; subsequently, the ST reactions were induced by the addition of both the Mn2+ and DNA substrate in (i), only the DNA substrate in (ii), and only Mn2+ in (iii). Aliquots were taken after 5, 10, 15, 20, 30, 40, and 50 min, and the product of the formation of the reactions versus the time plot were analyzed (Figure 5A). The result showed that Mn2+ strongly enhanced the ST activity of IN, with a signal 1.5 times that of IN pre-incubated alone. IN pre-incubated in the presence of DNA was almost the same active as IN pre-incubated alone. We subsequently performed the experiments in the presence of Mg2+ instead of Mn2+. The time plot curves of the Mg2+-dependent ST reactions were partially different from previous Mn2+-dependent reactions. Compared with IN pre-incubated alone, the pre-incubation of IN with Mg2+ also enhanced the ST activity notably, but not as significant as that of the Mn2+-dependent reaction. Unlike the Mn2+-dependent reaction in which the pre-incubation of IN with DNA had no effect on ST activity, the pre-incubation of IN with DNA enhanced the Mg2+-dependent ST activity remarkably (Figure 5B).
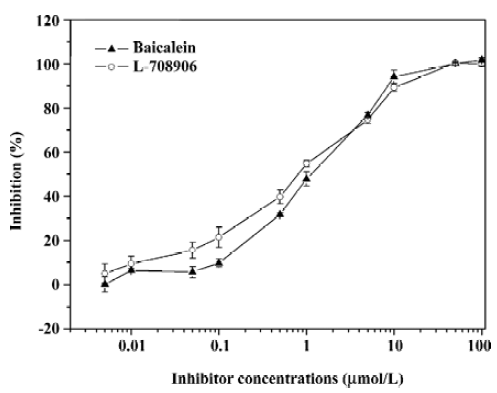
Antiviral compounds screening efficiency Baicalein and L-708906 were diluted in DMSO to a final concentration of 10% DMSO into the reaction volume and pre-incubated with IN at 37° C in the reaction buffer in the absence of Mn2+ for 10 min, followed by the addition of Mn2+ and the DNA substrates. The reactions were carried out at 37 °C for 1 h and the AP-coupled anti-DIG antibody and subsequent detection procedure was applied to detect the assay signals. The inhibition percentage and IC50 values were calculated based on the assay results using a non-linear regression curve fit. The IC50 values of baicalein and L-708906 for the IN ST reaction in this assay were 1.06 and 0.77 µmol/L, respectively (Figure 6).
Statistical evaluation of assay performance The screening window coefficient, Z (or Z') factor, has been defined to evaluate effective HTS assays[15]. It is capable of reflecting the assay signal dynamic range as well as the data variation associated with the signal measurements. In the present assay, all Z' factors were determined as described[15]:
Z'=1-[3×(SDPOS+SDNEG)/(MPOS-MNEG)]
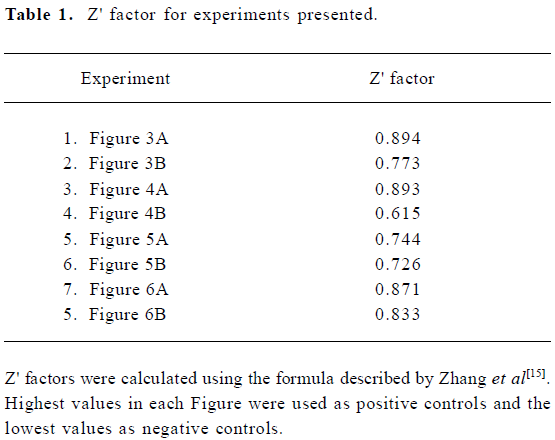
Calculations were based on the standard deviations (SD) and intensity means (M) of the controls. The highest values were used as the positive control and the no IN reactions (or no metal ion reactions) were used as the negative control. An analysis of the data resulting from the experiments present in these Figures showed that the assay we developed for IN ST activity had a Z' score ranging from 0.6 to 0.9 (Table 1). Together with the S/N and S/B ratios, these statistical parameters reflect the high sensitivity, specificity, and robustness of the assay.
Full table
Discussion
In the present study, we describe a novel assay that can be employed to measure the ST activity of IN as well as both the 3P and ST activities together. It is reported that ST reaction is the primary key to the effective suppression of viral replication, and compounds inhibiting the IN activity in vitro by interfering the 3P reaction alone lack antiviral activity[11,12]. Therefore, we applied this assay only in the ST reaction measurement in this work.
Several improvements have been acquired in this assay. First, the BIO-DIG combination was used and the assay was conducted in a 96-well microplate. This could be conducted in all kinds of multiwell microplate formats if desired, ensuring no radioactive waste and a high-throughput format. Second, the magnetic beads were used to capture the reaction product, therefore, all the reagents were freely suspended in solution instead of solid-phase assays in which donor or target DNA was attached to the microplate. Without the immobilization of DNA or the protein, each reagent can be added at any given time; it is more flexible to investigate the interaction of all reagents and it is easy to study the pharmacology of inhibitors targeting IN. Furthermore, with the application of magnetic beads, neither the precoating nor the blocking of microplate was required; it was less laborious and time consuming. After the last wash before the detection, the magnetic beads were easily transferred into fresh wells, and the background caused by the non-specific binding of DNA or the antibody was almost totally eliminated. Third, we applied this assay in 2 different detection systems. Both the AP conjugate and FITC conjugate anti-DIG antibodies and their following detection strategies were used. The result from both absorbance and fluorescence proves that the assay is specific and sensitive. Furthermore, the application of the fluorescent antibody needs no extra enzyme-coupled detection procedure, and the product formation is directly quantified by a fluorescent reader or flow cytometer. The entire assay can be accomplished in approximately 2 h; it is fast and convenient.
Divalent ion is necessary for both in vivo and in vitro activities of IN. In vitro assays use either Mg2+ or Mn2+ as the cofactor in reactions. Mg2+ is widely considered to be the biological relevant divalent cation cofactor for IN activity in vivo[16–19], but IN shows very low activity with Mg2+ as a cofactor, and Mn2+ is frequently used in most in vitro assays[8,10,20,21]. We optimized this assay in the presence of either Mg2+ or Mn2+ and found that the presence of Mn2+ instead of Mg2+ doubled the ST activity, but not as significantly as other reported works in which the Mn2+-dependent activity was approximately 4–6 times that of the Mg2+-dependent activity[13,20,21]. Using Mg2+ as the cofactor still showed a signal as high as 0.7 of A405. Lee et al reported that the 3P activity of IN alters depending on the structure and length of the oligonucleotide substrates, and increases in the length of the substrate cause alterations in the efficiency of Mg2+-dependent ST activity[22]. Hwang et al optimized their ST activity assay in the presence of Mg2+ using 35 bp donor DNA and suggested that the lengthening of donor DNA allowed Mg2+ as the cofactor[7]. It is clear that an increase in the length of the DNA substrate could enhance the Mg2+-dependent IN activities as suggested[7,22–24]. With the lengthening of donor DNA to 31 bp, the assay can be conducted in the presence of either cationic cofactor and extends the assay utility.
Previous in vitro studies for IN 3P reactions using Mg2+ as a cationic cofactor have reported that the pre-incubation of IN with DNA increases enzymatic activity, and IN-DNA interaction does not require a metal ion cofactor[14]. In contrast, Vink et al used Mn2+ in their study as the cationic cofactor and reported that the stable binding of IN to DNA requires Mn2+ and IN shows increased activity upon pre-incubation with Mn2+[25]. We observed complicated interactions between IN, DNA, and divalent ions in the ST reaction. When Mg2+ was used as the reaction cofactor, the pre-incubation of DNA with IN enhanced the ST activity remarkably, as compared with IN pre-incubated alone (Figure 5B). This indicates an IN-DNA binding step before ST reaction. The same observation has been reported and the DNA-binding step has been determined by steady-state fluorescence anisotropy[14,26]. However, when Mn2+ was used as the reaction cofactor, no difference was observed after pre-incubation of DNA with IN, as compared with IN pre-incubated alone (Figure 5B). The result indicates that the IN-DNA interaction requires Mn2+ when Mn2+ is involved as the cationic cofactor. As previously suggested by Vink et al, Mn2+ is necessary for the effective binding and correct positioning of DNA in the active site of IN[25]. Furthermore, when either Mn2+ or Mg2+ was used as the cofactor, pre-incubation of metal ion with IN increased the ST activity, but pre-incubation of IN with Mn2+ instead of Mg2+ changed the ST activity more significantly (Figure 5). This information indicates the existence of the metal ion-IN interaction and metal ion coordination step before the ST reaction, and the interaction is more important for the Mn2+-dependent ST reaction. Yi et al reported that the affinity of IN for DNA is increased in the presence of metal ions. The relative order of this effect is Mn2+>Mg2+, and it is suggested that the different metal ion effect may explain why Mn2+ is generally more effective than Mg2+ as a cofactor for catalysis in the in vitro assays[27]. Taken together, the information presented from our experiments indicates that the Mg2+-dependent and Mn2+-dependent ST activity is not equivalent. The difference of IN activities in the presence of either 2 cations has been reported and widely studied[23,25–30]. Engelman et al reported that IN displays more non-specific nuclease activity and less ionic strength in the presence of Mn2+ than in the presence of Mg2+ [23]. Moreover, it has been demonstrated that mutations located in the IN binding site significantly affect the Mg2+-dependent IN activity, but not the Mn2+-dependent activity[28], suggesting that the IN-DNA contact is quite different with these 2 cations and IN coordinates these 2 cofactors differently. The coordination property differences of Mg2+ and Mn2+ may result in different conformational changes of IN, therefore causing differences in the specificity of IN-DNA interactions and accounting for differences in the Mg2+-dependent and Mn2+-dependent IN activities as suggested[26,28,30]. It is confirmed that our assay can be adapted to study the ST reaction mechanism in a high-throughput format.
The assay we developed was designed to screen antiviral compounds targeting IN. Therefore, we tested the inhibitor screening efficiency of the microplate assay using 2 known integrase inhibitors: baicalein and 1 compound of the diketo acid family L-708906. The IC50 values are comparable to previous experiment data using other assays[8,20], indicating that this microplate assay is efficient and reliable in antiviral compound identification. This assay is optimized in multiwell microplate format, therefore has the potential of screening antiviral drug candidates in a high-throughput format.
In summary, the assay we presented here can be used for the rapid and specific detection of HIV-1 IN ST activity as well as for the efficient identification of drug candidates targeting IN. Furthermore, this assay can be also adapted to study the reaction character in a high-throughput manner. Future efforts will focus on using this assay to screen compound libraries to test HTS efficiency of the assay and identify new IN inhibitor candidates.
Acknowledgements
We thank Dr Li-ming HU, Department of Drug Synthesis, College of Life Science and Bioengineering, Beijing University of Technology, for providing the L-708906 compound.
References
- Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J Biol Chem 2001;276:213-16.
- Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem 2004;4:965-77.
- Pluymers W, De Clercq E, Debyser Z. HIV-1 integration as a target for antiretroviral therapy: a review. Curr Drug Targets Infect Disord 2001;1:133-49.
- Marchand C, Neamati N, Pommier Y. In vitro human immunodeficiency virus type 1 integrase assays. Methods Enzymol 2001;340:624-33.
- Chow SA. In vitro assays for activities of retroviral integrase. Methods 1997;12:306-17.
- Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res 1994;22:1121-22.
- Hwang Y, Rhodes D, Bushman F. Rapid microtiter assays for provirus topoisomerase, mammalian type IB topoisomerase and HIV-1 integrase: application to inhibitor isolation. Nucleic Acids Res 2000;28:4884-92.
- John S, Fletcher TM III, Jonsson CB. Development and application of a high-throughput screening assay for HIV-1 integrase enzyme activities. J Biomol Screen 2005;10:606-14.
- David CA, Middleton T, Montgomery D, Lim HB, Kati W, Molla A, et al. Microarray compounds screening (microARCS) to identify inhibitors of HIV integrase. J Biomol Screen 2002;7:259-66.
- Wang Y, Klock H, Yin H, Wolff K, Bieza K, Niswonger K, et al. Homogeneous high-throughput screening assays for HIV-1 integrase 3'-processing and strand transfer activities. J Biomol Screen 2005;10:456-62.
- Hazuda DJ, Felock PJ, Witmer M, Wolfe A, Stillmock K, Grobler JA, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 2000;287:646-50.
- Farnet CM, Wang B, Lipford JR, Bushman FD. Differential inhibition of HIV-1 preintegration complexes and purified integrase by small molecules. Proc Natl Acad Sci USA 1996;93:9742-7.
- He HQ, Ma XH, Liu B, Zhang XY, Chen WZ, Wang CX, et al. High-throughput real-time assay based on molecular beacons for HIV-1 integrase 3'-processing reaction. Acta Pharmacol Sin 2007;28:811-7.
- Smolov M, Gottikh M, Tashlitskii V, Korolev S, Demidyuk L. Kinetic study of the HIV-1 DNA 3'-end processing single-turnover property of integrase. FEBS J 2006;273:1137-51.
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67-73.
- Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochoon JC, et al. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry 2000;39:9285-94.
- Hindmarsh P, Lei J. Retroviral DNA integration. Microbiol Mol Biol Rev 1999;63:836-43.
- Sherman MP, Greene WC. Slipping through the door: HIV entry into the nucleus. Microbes Infect 2002;4:67-73.
- Andrake MD, Skalka AM. Retroviral integrase, putting the pieces together. J Biol Chem 1996; 271: 19 633–6.
- Tramontano E, Onidi L, Esposito F, Badas R, Colla PL. The use of a new in vitro reaction substrate reproducing both U3 and U5 regions of the HIV-1 3'-ends increases the correlation between the in vitro and in vivo effects of the HIV-1 integrase inhibitors. Biochem Phar 2004;67:1751-61.
- Debyser Z, Cherepanov P, Pluymers W, De Clercq E. Assays for the evaluation of HIV-1 integrase inhibitors. Methods Mol Biol 2001;160:139-55.
- Lee SP, Kim HG, Censullo ML, Han MK. Characterization of magnesium-dependent 3'-processing activity for human immunodeficiency virus type 1 integrase : real-time kinetic studies using fluorescence resonance energy transfer. Biochemistry 1995; 34: 10 205–14.
- Engelman A, Craigie R. Efficient magnesium dependent human immunodeficiency virus type 1 integrase activity. J Virol 1995;69:5908-11.
- Miller M, Bor YC, Bushman FD. Target DNA capture by HIV-1 integration complexes. Curr Biol 1995;5:1047-56.
- Vink C, Lutzke RAP, Plasterk RHA. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA. Nucleic Acids Res 1994;22:4103-10.
- Agapkina J, Smolov M, Barbe S, Zubin E, Zatsepin T, Deprez E, . Probing of HIV-1 integrase/DNA interactions using novel analogs of viral DNA. J Biol Chem 2006; 281: 11 530–40.
- Yi J, Asante-Appiah E, Skalka AM. Divalent cations stimulate preferential recognition of a viral DNA end by HIV-1 integrase. Biochemistry 1999;38:8458-68.
- Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein/DNA interaction. EMBO J 1998;17:5832-43.
- Diamond TL, Bushman FD. Role of metal ions in catalysis by HIV integrase analyzed using a quantitative PCR disintegration assay. Nucleic Acids Res 2006;34:6116-25.
- Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotrans-ferase enzymes. Proc Natl Acad Sci USA 2002;99:6661-6.
