Investigation of non-covalent complexes of glutathione with common amino acids by electrospray ionization mass spectrometry1
Introduction
Recently, research on characterizing the non-covalent complexes between small molecules and biomacromolecules, such as proteins, polypeptides, and oligonucleotides[1-5] has attracted much attention because of their potential application in pharmacology. As we know, in many cases, the biological function of a protein depends on its non-covalent interactions with other components existing in the living cell. Investigating these non-covalent interactions will be beneficial to unravel the mysteries of cellular function in health and disease states[6-7]. Therefore, it is urgent for us to explore the non-covalent interactions between small molecules and biomacromolecules.
Over the past decades, a variety of analytical techniques have been applied to determine ligand bindings, including spectrophotometry, fluorescence polarization, enzyme assay, and electrospray ionization mass spectrometry (ESI-MS). By virtue of its unrivaled speed, sensitivity, and low sample consumption, ESI-MS has become a powerful tool to probe the interactions of the protein hosts and the guests, including inhibitors, cofactors, metal ions, and nucleic acids. Furthermore, it can offer stoichiometric information of the complex directly and identify multiple components simultaneously, thus dramatic numbers of research articles have been reported[8-13]. For instance, Bligh et al[14] measured the dissociation constants of inhibitors non-covalently binding to the Src homology 2 (Src SH2) domain through non-linear regression curve fitting. Additionally, by using the sustained off-resonance irradiation collision-induced dissociation technique, Yu et al[15] continued their research on characterizing non-covalent complexes of (2,6-di-O-methyl)-β-cyclodextrin with active components from traditional Chinese medicines, including 3 quercetin O-glycosides, hyperoside, and rutin.
It is emphasized that the binding affinities of ligands to proteins measured by ESI-MS apply principally for the gas phase. So far, whether the gas phase constants can represent those in the solution phase remains controversial. Some of the most critical issues are as follows. First, in some cases, the ions arising from non-specific interactions are kinetically stable[16], as compared to the corresponding specific complexes during desolvation and ionization. Second, the ionic interactions are strengthened and hydrophobic interactions are destroyed when transferring in the gas phase[17]. Third, the signal suppression may become apparent at high analyte concentrations. Despite all this, many studies have demonstrated that the binding constants of macromolecular complexes[18] acquired by ESI-MS in the gas phase are found to correlate well to the measured affinity data derived from other solution-based methods.
As one of the most ubiquitous and important small biomolecules, glutathione (γ–glutamyl–cysteinyl–glycine) is present in the cells of all organisms at millimolar concentrations and possesses a multitude of physiological functions[19-22]. Reduced glutathione contains thiol (-SH), so it can be simplified as GSH. Glutathione is also one of the major cellular safeguards, which can offer triple protection as a key antioxidant, antitoxin, and cofactor. With the aid of γ-glutamyl transpeptidase, glutathione can actively transport the extracellular amino acids into cell through covalent binding in the Meister cycle[23]. Recently, the binding of phenylarsenic species to glutathione, isotocin, and thioredoxin by ESI-MS was quantitatively evaluated by Schmidt et al[24]. Their work prompted our renewed interest in exploring the non-covalent interaction between glutathione and other small molecular. To our knowledge, in human blood, the concentrations of glutathione[25] and common amino acids range from 1 to 3 mmol/L and from 1×10–2 to 1×10–6 mol/L, respectively, which makes it possible to form the non-covalent complexes of glutathione with amino acids in such circumstances. However, the report on the non-covalently binding of glutathione to common amino acids is still deficient.
In this work, using the ESI-MS technique, we investigated the interaction between glutathione and common amino acids in the equilibrium system in vitro and characterized their non-covalent complexes. The binding of the complexes was further examined by collision-induced dissociation in a tandem mass spectrometer as well as UV spectroscopy. To avoid distinct ionization efficiency discrepancy and signal suppression in the ESI-MS measurements, the interactions between GSH and Glu were quantitatively evaluated. Due to the existence of some oligomeric species, an improved formula was derived to calculate the dissociation constants of the complexes. Finally, a possible formation mechanism for the complexes of glutathione with amino acids was discussed.
Materials and methods
Chemicals Glutathione, purchased from Sigma (St Louis, MO, USA), was used without further purification. The common amino acids were obtained from Shanghai Bio-Basic (Shanghai, China). Methanol was obtained from Sino-Pharmaceutical Chemical (Shanghai, China). All chemicals were dissolved in deionized water and prepared into 1.0×10–3 mol/L stock solutions.
Instruments A triple–quadrupole mass spectrometer (API III, PE-Sciex, Concord, ON, Canada, m/z 2400) equipped with a SP101i Syringe Pump (World Precision Instruments, Sarasota, FL, USA) and a Sciex ionspray interface (Sciex, Concord, ON, Canada) was used to generate multiply charged ions by spraying the sample solution through a stainless steel capillary. The voltage on the sprayer was set at 4800 V for positive ion production. The mass scale of the spectrometer was calibrated with polypropylene glycol. Nitrogen was used for nebulizer gas and curtain gas.
The CID experiments were performed on a hybrid ion trap and time-of-flight mass spectrometer with NanoESI interface (Shimadzu, Kyoto, Japan). The positive ESI conditions were as follows: high voltage probe, 1.5 kV; skimmer voltage, 10 V; syringe pump flow, 200 nL/min; and ion source temperature, 200 °C. CID parameters were chosen: 50% for CID energy and 50% for collision gas parameter. Argon was used for cooling and collision gas. CID collision time was 30 ms. Detector voltage of time-of-flight was 1.7 kV. A solution of CF3COONa was used as the standard sample to adjust resolution and to perform mass number calibration.
UV spectroscopy was detected in a Shimadzu UV-2401 UV spectrophotometer (Japan) at room temperature. The concentration of the His, Phe, Trp, and GSH solutions used in the spectroscopic measurements was the same as 0.1 mmol/L. The concentration of the incubated (30 min at 21°C) binary solution of His, Phe, or Trp, and GSH was also 0.1 mmol/L. The scan range was 190-230 nm.
Procedures For each system, a series of samples with the constant concentration of glutathione and different concentrations of amino acids were mixed without adding acid or base. After mixing evenly, the mixture was then incubated at a room temperature of 21°C for 30 min to bring into the state of equilibrium. To increase the intensity of ion peak in the mass spectra, 5% methanol was added before the ESI-MS determination. All samples were operated in positive polarity mode.
Calculation formula A previous study[24] demonstrated that some factors, such as ionization efficiency and signal suppression could strongly influence the accuracy of the dissociation constants of complex in the ESI-MS measurements. Therefore, it is necessary to quantitatively evaluate the ionization efficiency of free glutathione and free amino acids by measuring concentration series of glutathione and amino acids separately, and comparing the ion intensities obtained for same concentrations.
In these cases, 3 assumptions can be made. First, no change occurs when the mixture is transferred from the solution phase to the gas phase, and no dissociation reaction occurred in the complexes. Second, the measured peak intensities of free glutathione, free ligand, the complexes, and oligomers are directly proportional to their equilibrium concentrations in the solution. Third, free glutathione, free ligand, complexes, and oligomers share similar ionization efficiency.
One-ligand system According to the assumptions mentioned earlier, the calculation method of Kd is derived. Due to the presence of some non-specific oligomeric complexes arising from glutathione or amino acids, the formula for dissociation constants of complexes was obtained by improving the published formula[26]. The polypeptide glutathione and common amino acid ligands are simplified as P and L in our article, respectively. For a system containing 1 kind of amino acid binding to glutathione with one or many binding sites, the dissociation constants are defined as:
PL1 Δ P+L Kd,1
PL2 Δ P+2 L Kd,2
PLn Δ P+n L Kd,n
P2 Δ P+P KP,1
P3 Δ P+2 P KP,2
Pi +1 Δ P+i P KP,i
L2 Δ L+L KL,1
L3 Δ L+2 L K L,2
Lj +1 Δ L+ j L KL,j
(P), (L), and (PLn) represent the equilibrium concentration of P (glutathione), L (ligand), and PLn (the complex of glutathione with ligand), respectively, in which n represents the number of binding sites. In the following text, (Pi+1) and (Lj+1) represent the equilibrium concentration of Pi+1 and Lj+1, where i+1 and j+1 represent the monomer numbers of glutathione and ligand, respectively.
We proposed that (P)0 is the total concentration of glutathione in the solutions, which included glutathione, the complexes of glutathione with ligands, and self-polymers of glutathione: (P)0=(P)+(PL)+(PL2)+ ······ +(PLn)+2(P2)+3(P3)+ ······ +(i+1) (Pi+1), where (L)0 is the total concentration of amino acid ligands in the solutions, which included amino acid ligands, the complexes of glutathione with ligands, and self-polymers of ligands, (L)0=(L)+(PL)+2(PL2)+ ··· + n (PLn)+2 (L2) +3(L3)+ ··· +(j+1) (Lj+1). Consequently, the relative abundances obtained from mass spectral data are reflected in the concentrations in the solution phase, Σ represented the relative ion intensity of different species: (P)0∝ total intensities of ΣP, ΣPL1··· ΣPLn, and 2ΣP2, 3ΣP3 ··· (i+1)ΣPi+1; (L)0∝ total intensities of ΣL, ΣPL1, 2ΣPL2··· nΣPLn, and 2ΣL2, 3ΣL3···(j+1)ΣLj+1; (PLn)∝ total intensities of ΣPLn.
The intensities of P(ΣP), PL1 (ΣPL1) ··· PLn (ΣPLn), and P2(ΣP2), P3(ΣP3), ···
Pi+1(ΣPi+1), and L(ΣL), L2(ΣL2), L3(ΣL3) 贩? Lj+1 (ΣLj+1) are easily determined by ESI-MS and the values of an (a1,a2, ···, an), bi (b1,b2···bi), cj (c1,c2···cj) can be calculated as follows:
(PL1)=a1 (P)0 (13)
(PL2)=a2 (P)0 (14)
(PLn)=an (P)0 (15)
(P) = (P)0?([PL]+[PL2]+ ··· +[PLn]+2[P2]+3[P3]+ ··· +[i+1][Pi+1])
=(P)0?(a1+a2+ ··· +an+2b1+3b2+ ··· +[i+1]bi) (P)0
=(1朳a1+a2+ ··· +an+2b1+3b2+ ··· +[i+1]bi]) (P)0 (16)
=
(L)=(L)0?([PL]+2[PL2]+ ··· +n[PLn]+2[L2]+3 [L3]+ ··· +[j+1][Lj+1])
=(L)0?(a1+2a2+ ··· +n an) (P)0?(2c1+3c2+ ··· +[j+1]cj) (L)0 (17)
=
Put simply, in equation 20, Kd,n can be represented as a general equation:
According to equation 21, the Kd,m of PLm (1) can be obtained for a system containing 1 kind of ligand.
Results
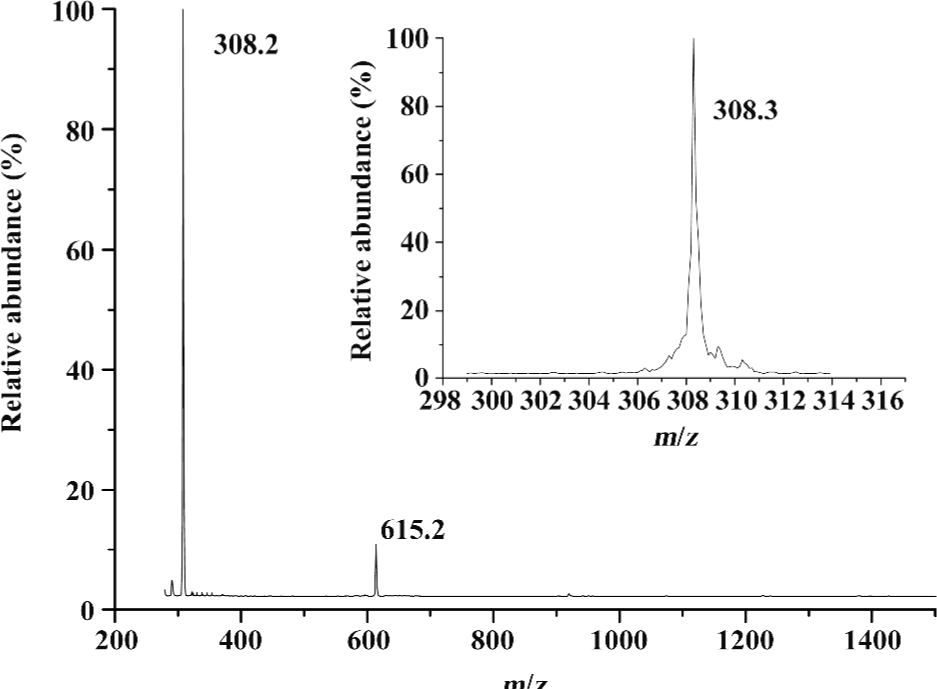
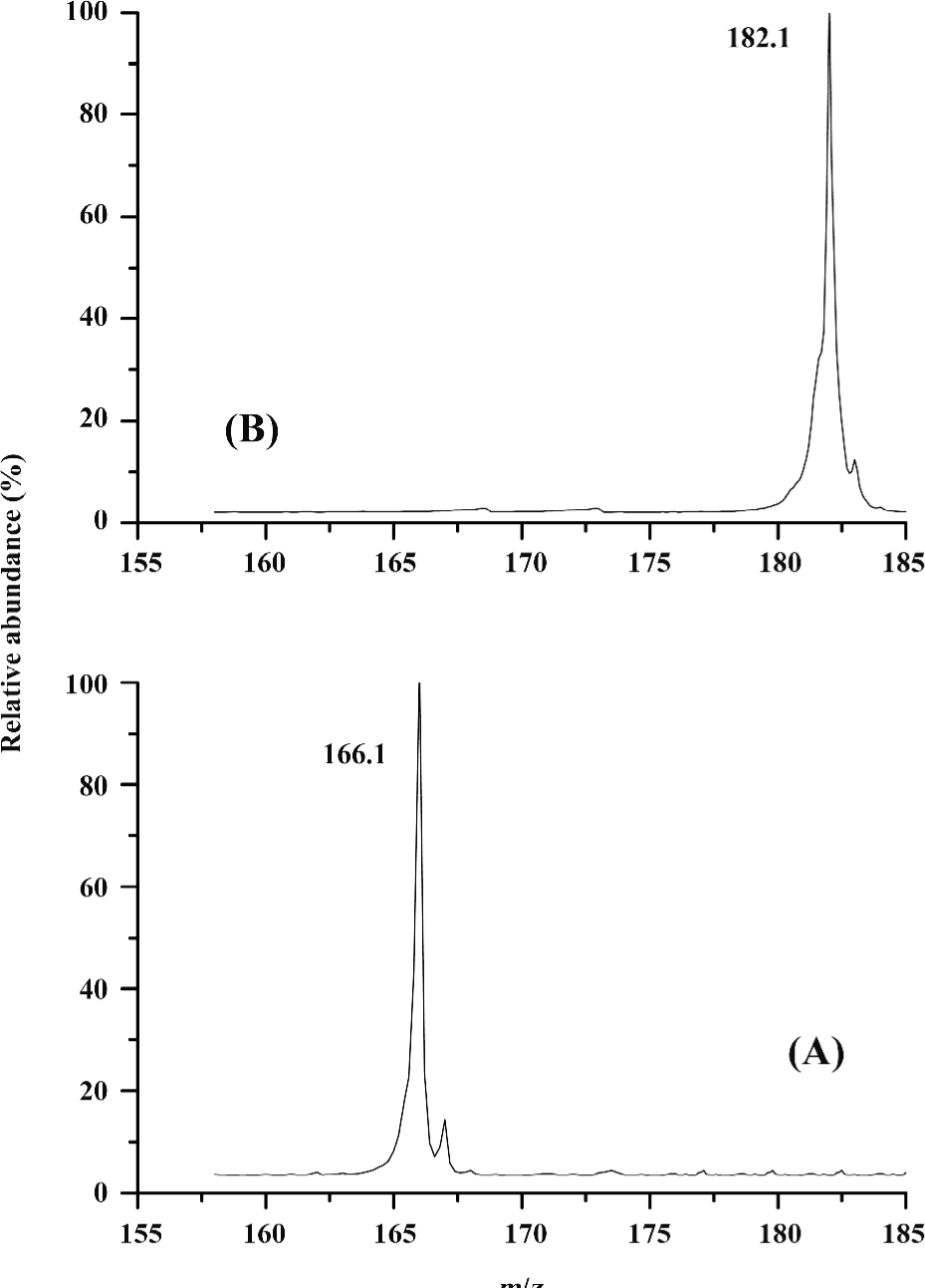
ESI mass spectra of several amino acids and glutathione Glutathione is a tripeptide consisting of Glu, Cys, and Gly. The molecular mass of glutathione is 307.3. Ions of (P+H)+ for glutathione were in great abundance. The peaks at m/z 308.3 and 615.2 represented those of (P+H)+ and (P-P+H)+ dimers, respectively (Figure 1). The non-covalently-bound glutathione dimer could be such a non-specific complex formed during the ESI process. It is noted that the non-covalently-bound glutathione dimer was different from the oxidized glutathione (GSSG), in which the thiol (-SH) group was oxidized, resulting in the elimination of the hydrogen of the thiol. It is inferred that the non-covalently-bound GSH dimer can still retain its reducing ability to deal with the oxidation stress. The ESI-MS for two 2 typical amino acids, Phe and Tyr, were measured, and the prominent molecular ion peaks at m/z 166.1 and 182.1 could be attributed to (Phe + H)+ and (Tyr + H)+, respectively (Figure 2). It can be concluded that the single-charged state ions for the amino acids could be produced easily in the ESI-MS.
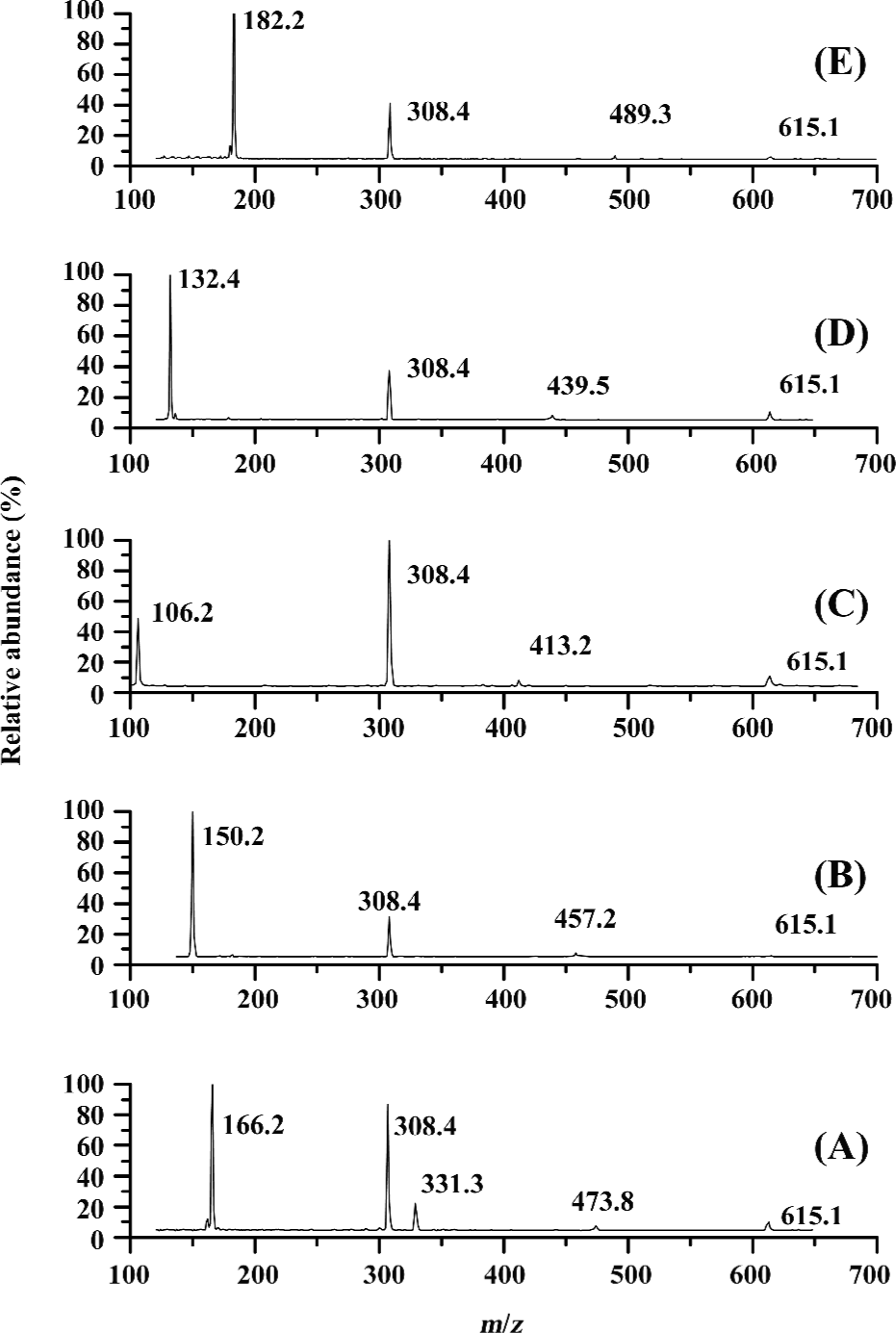
ESI mass spectra for the complexes of glutathione with Phe, Met, Tyr, Ser, and Ile ligands The ESI mass spectra for the complexes of glutathione with Phe, Met, Tyr, Ser, and Ile indicate that with the exception of amino acid, glutathione, its complex with 1 ligand (PL), and dimer of glutathione or Phe, the peaks for the complexes with 2 or more ligands have not been identified (Figure 3). For the glutathione and Phe systems, the m/z of 473.2, 331.3, 615.1 were attributed to the peaks for (P-L+H)+, (L-L+H)+, and (P-P+H)+, respectively (Figure 3A). It is recognized that in this system, the self-aggregation reaction of glutathione and Phe could take place practically, resulting in the formation of dimeric species. However, such phenomena did not occur in the other 4 systems. In contrast, the peaks of the m/z of 457.2, 413.2, 439.5, and 489.2 indicated the formation of the non-covalent complexes of (P-Met+H)+, (P-Ser+H)+, (P-Ile+H)+, and (P-Tyr+H)+, respectively (Figure 3B-3E).
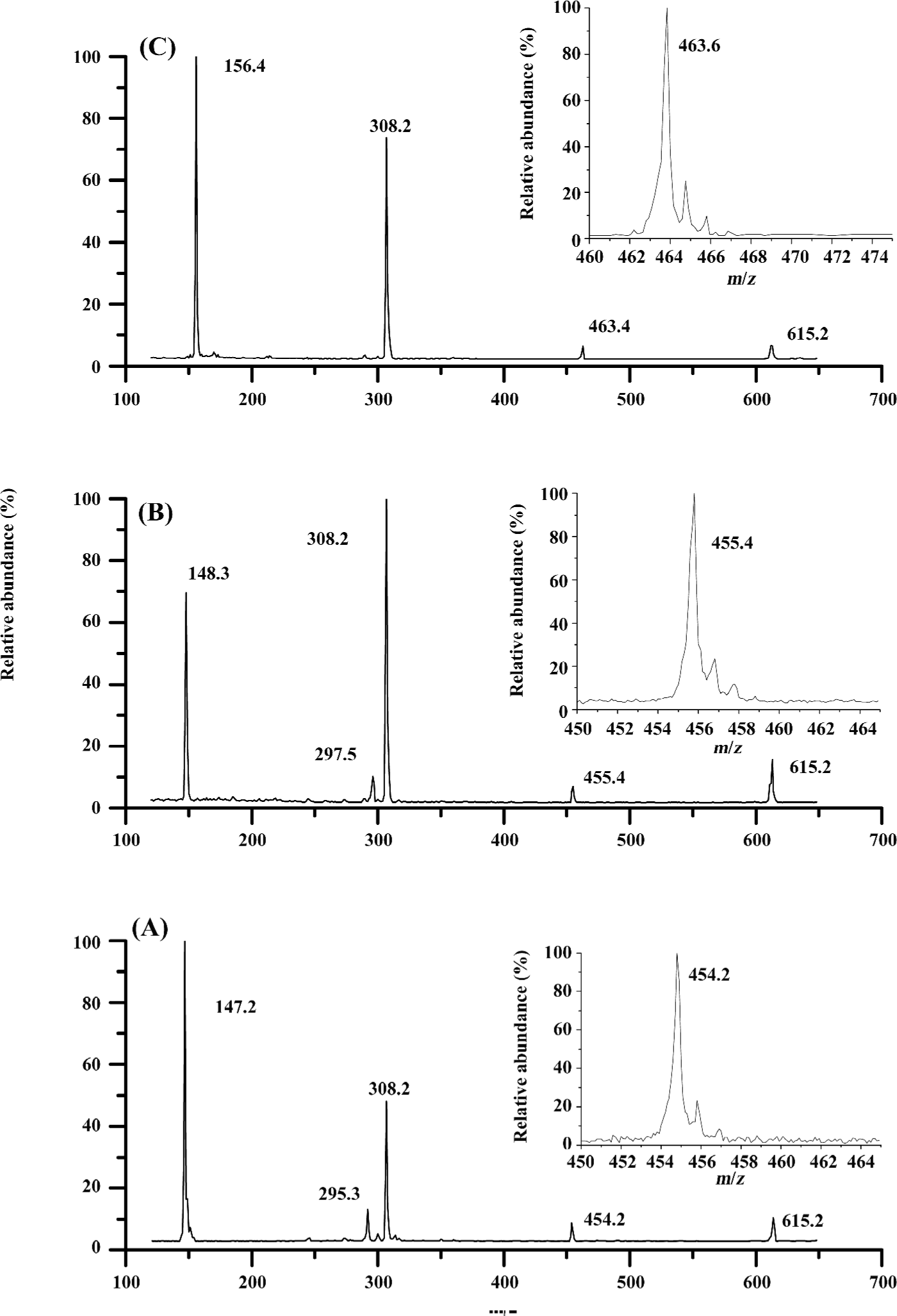
ESI mass spectra for the complexes of glutathione with Glu, His, and Gln The 3 amino acids, Gln, Glu, and His, are representatives for neutral, acidic, and basic amino acids, respectively (Figure 4). The m/z of 454.2, 455.4, 463.6 were attributed to the ion peaks for (P-Gln+H)+, (P-Glu+H)+, and (P-His+H)+, which indicated that the non-covalent complexes of GSH with Gln, Glu, or His could form in their mixed solutions (Figure 4A-4C). In addition, the monoprotonated dimeric species, such as (Gln-Gln+H)+ and (Glu-Glu+H)+ also appeared in m/z 295.3 and 297.5, respectively (Figure 4A,4B). To further investigate the binding affinities of ligands to glutathione, the dissociation constants of these complexes were calculated in Tables 1-3.
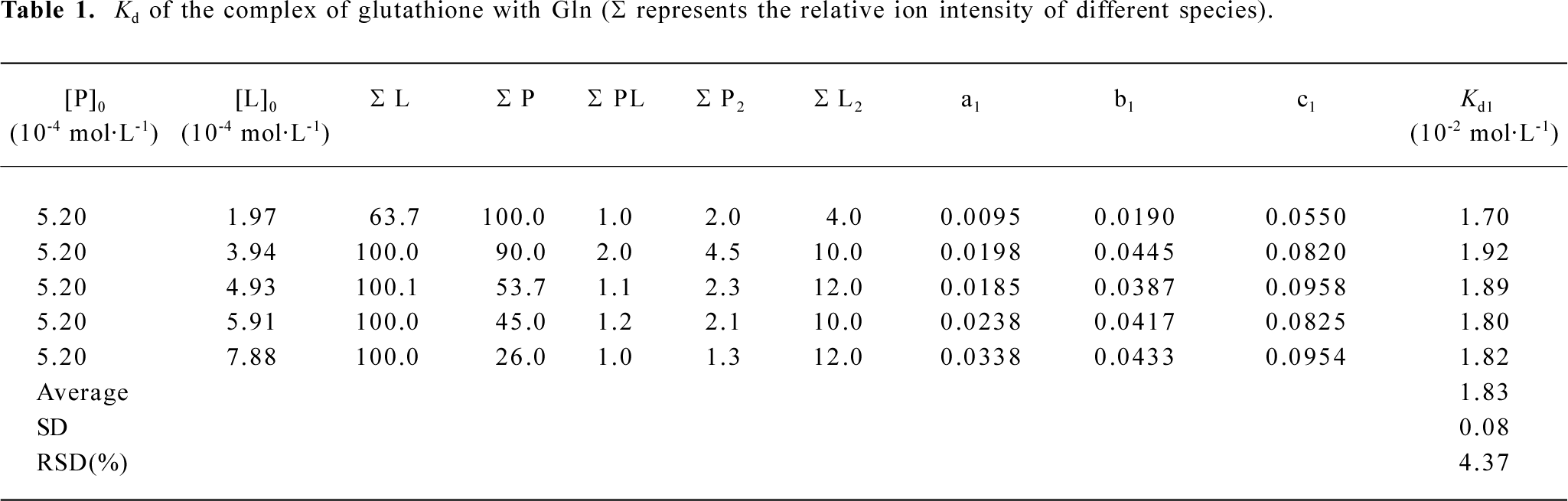
Full table
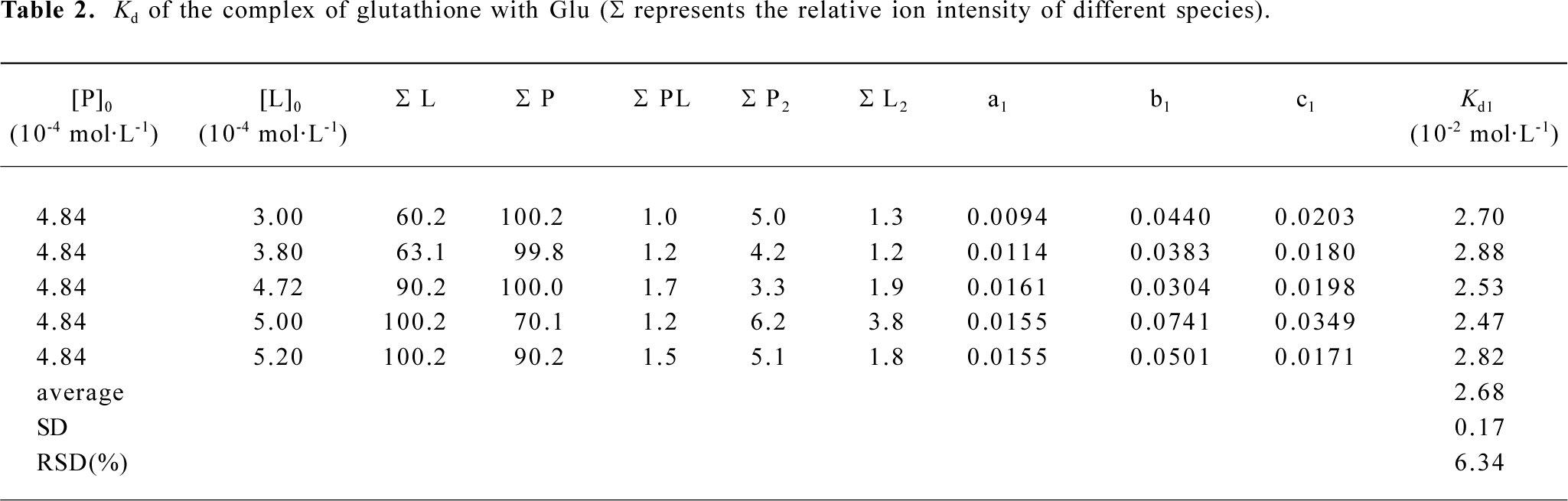
Full table
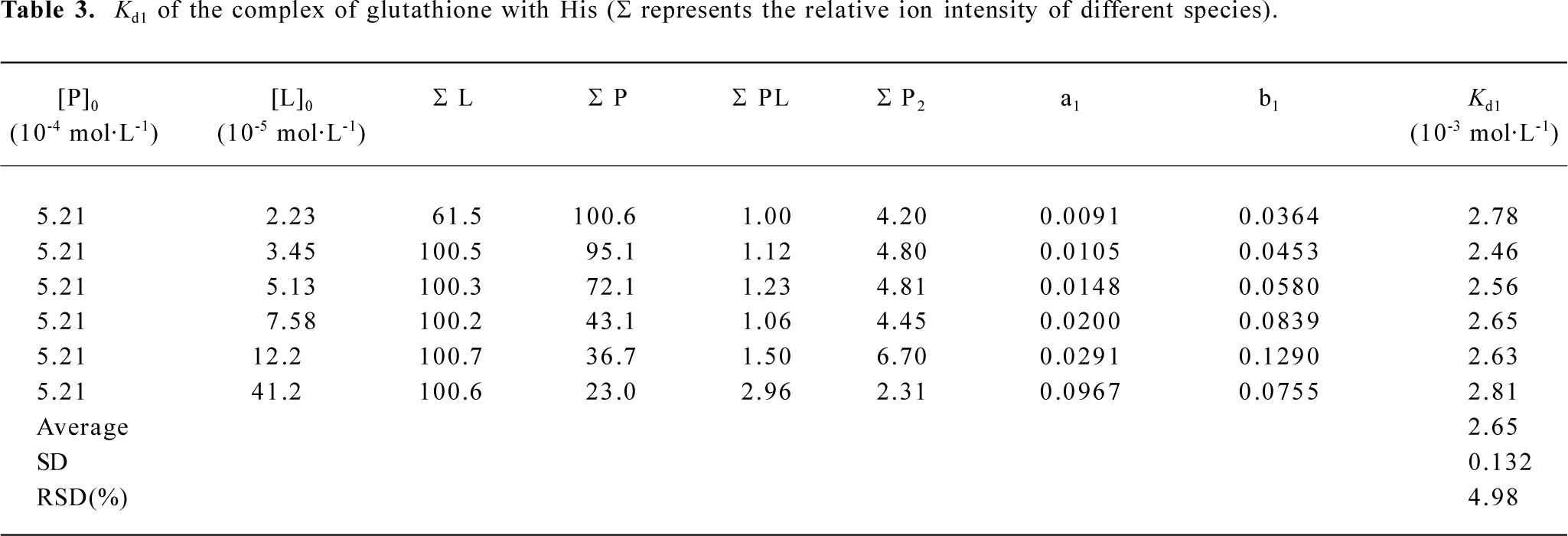
Full table
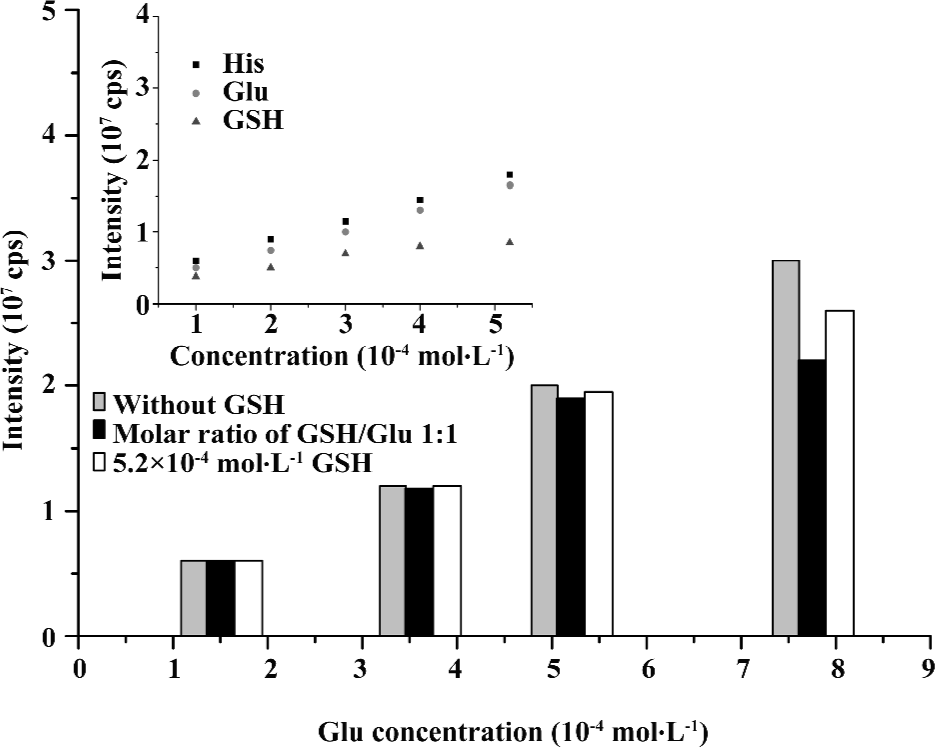
Quantitative evaluation of interactions of glutathione with Glu The dependence of the ESI-MS signal intensity for (M+H)+ of GSH (m/z 308.3), Glu (m/z 148.1), or His(m/z 156.2) on concentrations was examined (Figure 5). The curve of GSH exhibited a saturation characteristic, whereas those of 2 amino acids rose up monotonously. This was indicated by increasing concentrations; His shared similar ionization efficiency with Glu, whereas that of GSH was slightly lower. The suppression effect was not apparent in case of the addition of GSH to Glu when the Glu concentration was lower than 5.2×10–4 mol/L. However, a minor signal suppression began to emerge when the concentration of Glu extended to 5.2×10–4 mol/L. Despite this problem, the decrease of signal intensities for Glu at a fixed concentration of GSH (5.2×10–4 mol/L) was less than those for Glu at the same molarities of GSH and Glu (Figure 5).
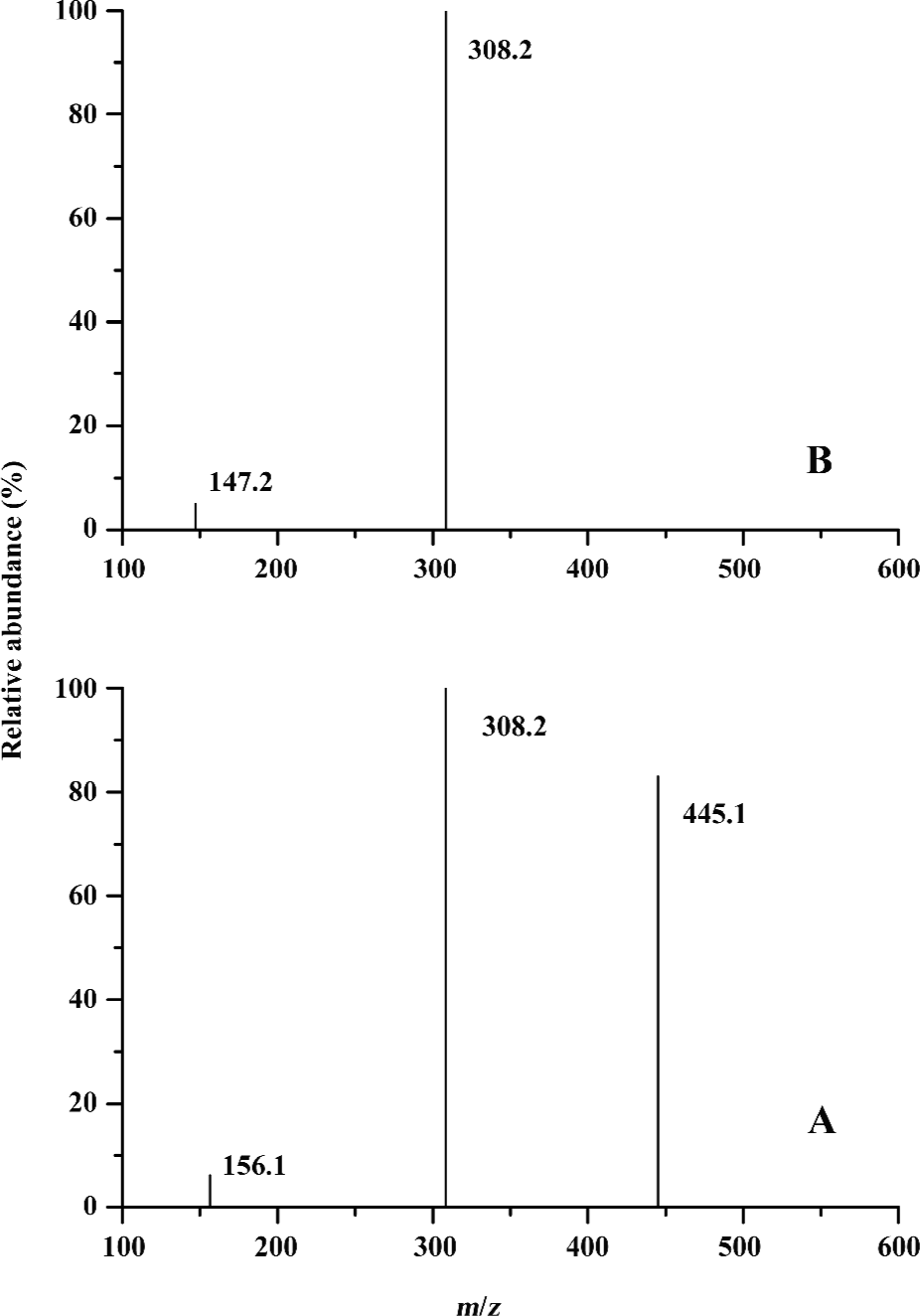
CID spectra of non-covalent complexes of glutathione with His or Gln To further validate the non-covalent binding of glutathione to amino acids, CID experiments on the precursor ion (GSH+L+H)+ were performed, where L represented His or Gln (Figure 6). The fragment ions at approximately m/z 156.2 and 308.2 arising from the precursor ion (m/z 463.2) were attributed to (M+H)+ for His and GSH, respectively (Figure 6A). The fragment ion (m/z 445.1) arising from losing neutral H2O was also observed. Similarly, for the precursor ion (m/z 454.2), the fragment ions of [M+H]+ for Gln (m/z 147.2) and GSH (m/z 308.2) were also identified (Figure 6B). Combining CID evidence with data from Figure 4, it can be concluded that amino acids can interact with glutathione to form non-covalent complexes.
UV spectroscopy Recently, the configuration of arsenite lutathione complexes was determined by Han et al using electrochemical scanning tunneling microscopy and found the binding of glutathione to arsenite in solution could also be confirmed by UV spectroscopy[27]. Moreover, the molar extinction coefficients of common amino acids of UV absorption at 214 nm have been also measured[28], and the results indicated that His, Phe, and Trp were strong contributors to the absorbance at 214 nm.
We determined the UV spectroscopy ranging from 190 to 230 nm for GSH, Phe, His, and Trp together with 3 incubated GSH–amino acid binary solutions. It was observed that the absorption peaks of 211, 208, and 218nm for His, Phe, and Trp were shifted to 213, 210, and 220 nm for His– GSH, Phe–GSH, and Trp–GSH binary solutions, respectively. Particularly, an obvious 192 nm absorption peak was observed in the Phe–GSH binary solution. The minor shifts of UV spectra validated the formation of the GSH–His, Phe, or Trp complexes
Calculation of Kd for the complexes of glutathione with Gln, Glu, and His ligands According to the results of Figure 5, the concentration of either glutathione or amino acids chosen for the dissociation constant measurements by ESI-MS should be lower than 5.2×10–4 mol/L so that the distinct ionization efficiency discrepancy and signal suppression effect could be avoided. Moreover, the fixed concentration of GSH was chosen in the dissociation constants measurement below.
The relative abundances for all ion peaks were obtained from the mass spectra data. According to equation 21, the dissociation constants? Kd values of the complexes of glutathione with Gln, Glu, and His could be obtained by calculating the an, bi, and cj values (Tables 1-3). For the complexes of Gln, Glu, and His, the relative standard deviations (RSD) of their dissociation constants were 4.37, 6.34, and 4.98, respectively, which were satisfying. Because the His– His dimer was not identified as Glu in ESI-MS, cj could be omitted in the calculation (Table 3). The Kd1 (1×10–2 mol/L) for the complex of glutathione with Glu was larger than that (1×10–3 mol/L) for the complex of glutathione with His, indicating that the affinity between glutathione and Glu was comparatively weaker (Tables 2, 3).
Curve fitting For the system of 1 ligand interacting with glutathione, the Kd,1 values can also be calculated approximately with the non-linear least squares regression method according to equation 22 in the literature[14]. For convenience, the products of self-polymer of P and L, such as P2, L2 are not considered in this formula. The curve fitting calculation formula is written as:
In equation 22 formula, y can be calculated from the intensities of PLn+ and P n+ of equation 23.
In the process of curve fitting, (P)0 and (L)0 are chosen as independent variables while y as dependent variables. By comparison, it can be seen that the Kd,1 values obtained from curve fitting without considering oligomeric species were slightly different from those obtained from our improved calculation formula in this study (Table 4). For instance, in the case of the Gln complex with glutathione, the Kd,1 (2.05×10–2) obtained from curve fitting was larger than that (1.83×10–2) from our improved formula. Therefore, it is recognized for the complexes of 1 ligand binding to glutathione, the Kd,1 values calculated accurately from our improved formula should be more reasonable to reflect the extent of complex formation.
Full table
Discussion
In the present study, we made an attempt to investigate the interaction of glutathione and each of 8 common amino acids in vitro by ESI-MS techniques, and found that glutathione and the 8 typical amino acids could form stoichiometric non-covalent complexes. The binding of the complexes was further confirmed by CID experiments in a tandem mass spectrometer as well as UV spectroscopy. The experimental results showed that only the complex of glutathione with 1 ligand was identified. Alternatively, glutathione only provided 1 binding site for amino acids to bind. Previous reports demonstrated that the complex of the Src SH2 domain with its inhibitors[14] can form 2 or more binding sites. Compared to the macromolecules of the Src SH2 domain, we think the relatively small size may limit the binding site number offered by glutathione.
Despite the non-covalent binding of glutathione to amino acids, the biological active site thiol in Cys of glutathione (γ- Glu–Cys–Gly) has not been affected yet, that is, the complexes can still retain their reducing ability to cope with the oxidation challenge in living system. This is primarily because no oxidation-reduction reaction occurred in the formation process of the non-convalent complexes. These complexes may influence their pharmacokinetics in blood to some extent.
In general, the dissociation constants determined by ESI-MS are valid, principally for the gas phase. It is better to investigate these equilibria additionally with other solution-based techniques, such as potentiometric titrations, nuclear magnetic resonance, or UV-vis spectrophotometry. When applying ESI-MS to measure the dissociation constants of complexes in solutions, some critical issues must be considered. For example, the correlation between the ionization efficiencies of different species needs to be evaluated (Figure 5). Amino acids, such as His and Glu shared similar ionization efficiencies in a wide concentration range. However, with the increasing of the concentrations, the ionization efficiency of GSH became lower than those of amino acids. Therefore, appropriate concentrations should be chosen for ESI-MS measurements.
Under some circumstances, there are complexes formed in solutions that are not stable during ESI or that show low gas phase stability. Once this situation really occurs, ionization efficiency of the complex will be influenced. If more serious, even the signal of the complexes can not be identified, so it is better to evaluate ionization efficiency of complex simultaneously. However, for the GSH complex in this work, its yield is comparatively lower, the formation mechanism as well as the participating atom is unknown, so it is difficult to evaluate the ionization efficiency of the complex directly.
To establish the calculation formula for the dissociation constants of non-convalent complexes, we quantitatively evaluated the interactions of Glu with glutathione. Consequently, distinct ionization efficiency discrepancy and signal suppression effect were avoided in our ESI-MS measurements. Our formula indicated that if the initial concentrations of glutathione and amino acids had been known, the dissociation constants can be obtained directly according to the peak intensities data offered by ESI-MS instead of curve fitting. It should be noted that our calculation formula can be also applied to other non-covalent complex systems containing oligomeric species.
The formation of the non-covalent complexes was confirmed by CID experiments (Figure 6). As the stability of the non-covalent bonds was much weaker than other covalent ones, the dissociation of non-covalent bonds was easy in the CID, which led to the fragment ions arising from GSH and amino acids. Since the His or Gln-bound fragment ions have not been identified, the binding site information cannot be provided directly. More work, such as Fourier transform ion cyclotron mass spectrometry[15] needs to be done to provide more structural information. In any case, based on the discussion mentioned earlier, the possible mechanism of glutathione binding to amino acids can still be proposed. First, the amino acids may approach the thiol (–SH) group of Cys or N-terminal amino group of Glu residue in glutathione through electrostatic attraction force; second, the hydrogen bond action between the amino acids and 1 group of glutathione would lead to the formation of the non-convalent complexes. The group of glutathione binding to the amino acid could be either thiol group of Cys or the amino group of Glu residue. The formation of the hydrogen bond between amino acids and proteins, such as cytochrome c was also found in other previous work[11].
In conclusion, the successful identification of ion peaks for the complexes of glutathione with each of the 8 common amino acids by ESI-MS in vitro revealed that glutathione could non-covalently bind to amino acids in human blood. The binding of the complexes was further confirmed by CID experiments in a tandem mass spectrometer as well as UV spectroscopy. In 1 ligand system, the dissociation constants for the complexes of glutathione with Glu, Gln, and His were quantitatively determined by an improved calculation formula. Based on our investigation, it can be inferred that the amino acids or glutathione may non-covalently bind to other biomolecules, such as proteins or oligonucleotides in the living system, respectively.
Author contribution
Yan-qiu CHU designed research; Zhao-yun DAI performed research; Chuan-fan DING contributed new reagents or analytic tools; Bo WU and Liang WU analyzed data; Chuan-fan Ding and Yan-qiu CHU wrote the paper.
References
- Konig S, Hasche A, Pallast S, Krieglstein J, Klumpp S. Detection of ATP-binding to growth factors. J Am Soc Mass Spectrom 2008;19:91-5.
- Yanes O, Aviles FX, Roepstorff P, Nazabal A, Zenobi R, Calvete JJ. Exploring the 搃ntensity fading? phenomenon in the study of non-covalent interactions by MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom 2007;18:359-67.
- Fabris D, Chaudhari P, Hagan N, Turner K. Functional investigations of retroviral protein-ribonucleic acid complexes by nanospray Fourier transform ion cyclotron resonance mass spectrometry. Eur J Mass Spectrom 2007;13:29-33.
- Wilson JJ, Brodbelt JS. Infrared multiphoton dissociation of duplex DNA/drug complexes in a quadrupole ion trap. Anal Chem 2007;79:2067-77.
- Baytekin B, Baytekin HT, Schalley CA. Mass spectrometric studies of non-covalent compounds: why supramolecular chemistry in the gas phase? Organ Biomol Chem 2006;4:2825-41.
- Bovet C, Wortmann A, Eiler S, Granger F, Ruff M, Gerrits B, et al. Estrogen receptor-ligand complexes measured by chip-based nanoelectrospray mass spectrometry: An approach for the screening of endocrine disruptors. Protein Sci 2007;16:938-46.
- Lin ZPJ, Li WK, Dai GW. Application of LC-MS for quantitative analysis and metabolite identification of therapeutic oligonucleotides. J Pharmaceut Biomed Anal 2007;44:330-41.
- Li MH, Yin LL, Cai MJ, Zhang WY, Huang Y, Wang X, et al. Design, synthesis, and anti-inflammatory evaluation of a series of novel amino acid-binding 1,5-diarylpvrazole derivatives. Acta Pharmacol Sin 2005;26:865-72.
- Terrier P, Tortajada J, Buchmann W. A study of non-covalent complexes involving single-stranded DNA and polybasic compounds using nanospray mass spectrometry. J Am Soc Mass Spectrom 2007;18:346-58.
- Mazzitelli CL, Brodbelt JS. Probing ligand binding to duplex DNA using KMnO4 reactions and electrospray ionization tandem mass spectrometry. Anal Chem 2007;79:4636-47.
- Lu HJ, Guo YL, Yang PY. Using amino acids for probing structure information of cytochrome c by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 2004;15:1612-5.
- Lear BJ, Kubiak CP. Origins of cooperative non-covalent host-guest chemistry in mixed valence complexes. J Phys Chem B 2007;111:6766-71.
- Li Y, Zhong DF, Chen SW, Maeba I. Identification of some benproperine metabolites in humans and investigation of their antitussive effect. Acta Pharmacol Sin 2005;26:1519-26.
- Bligh SWA, Haley T, Lowe PN. Measurement of dissociation constants of inhibition binding to Src-2 SH2 domain protein by non-covalent electrospray ionization mass spectrometry. J Mol Recognit 2003;16:139-47.
- Yu Z, Cui M, Yan CY, Song FR, Liu ZQ, Liu SY. Investigation of heptakis (2,6-di-O-methyl)-beta-cyclodextrin inclusion complexes with flavonoid glycosides by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 2007;21:683-90.
- Wang W, Kitova EN, Klassen JS. Bioactive recognition sites may not be energetically preferred in protein-carbohydrate complexes in the gas phase. J Am Chem Soc 2003; 125: 13 630-1.
- Wigger M, Eyler JR, Benner SA, Li WQ, Marshall AG. Fourier transform-ion cyclotron resonance mass spectrometric resolution, identification, and screening of non-covalent complexes of Hck Src homology 2 domain receptor and ligands from a 324-member peptide combinatorial library. J Am Soc Mass Spectrom 2002;13:1162-9.
- Nyadong L, Green MD, De Jesus VR, Newton PN, Fernandez FM. Reactive desorption electrospray ionization linear ion trap mass spectrometry of latest-generation counterfeit antimalarials via non-covalent complex formation. Anal Chem 2007;79:2150-7.
- Baydas G, Sonkaya E, Tuzcu M, Yasar A, Donder E. Novel role for gabapentin in neuroprotection of central nervous system in streptozotocine-induced diabetic rats. Acta Pharmacol Sin 2005;26:417-22.
- Levtchenko E, De Graaf-Hess A, Wilmer M, Van den Heuvel L, Monnens L, Blom H. Altered status of glutathione and its metabolites in cystinotic cells. Nephrol Dial Transplant 2005;20:1828-32.
- Sheng R, Gu ZL, Xie ML, Zhou WX, Guo CY. EGCG inhibits cardiomyocyte apoptosis in pressure overload-induced cardiac hypertrophy and protects cardiomyocytes from oxidative stress in rats. Acta Pharmacol Sin 2007;28:191-201.
- Chin TY, Chueh SH, Tao PL. S-nitrosoglutathione and glutathione act as NMDA receptor agonists in cultured hippocampal neurons. Acta Pharmacol Sin 2006;27:853-60.
- Njalssson R, Norgren S. Physiological and pathological aspects of GSH metabolism. Acta Paediat 2005;94:132-7.
- Schmidt A C, Neustadt M, Otto M. Quantitative evaluation of the binding of phenylarsenic species to glutathione, isotocin, and thioredoxin by means of electrospray ionization time-of-flight mass spectrometry. J Mass Spectrom 2007;42:771-80.
- Yao X, Wang YT, Chen G. Simultaneous determination of aminothiols, ascorbic acid and uric acid in biological samples by capillary electrophoresis with electrochemical detection. Biomed Chromatogr 2007;21:520-6.
- Zhang HR, Chen G, Wang L, Ding L, Tian Y, Jin WJ, et al. Study on the inclusion complex of cyclodexin and sulphonated azo dyes by electrospray ionization mass spectrometry. Int J Mass Spectrom 2006;252:1-10.
- Han MJ, Meng XG, Lippincott L. Determination of configuration of arsenite-glutathione complexes using ECSTM. Toxicol Lett 2007;175:57-63.
- Kuipers BJH, Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J Agric Food Chem 2007;55:5445-51.
