Overexpression of coxsackie and adenovirus receptor inhibit growth of human bladder cancer cell in vitro and in vivo1
Introduction
Coxsackie and adenovirus receptor (CAR) has primarily been studied for its role as the initial cell surface attachment receptor for coxsackie and group C adenoviruses. CAR is a 46 kDa class I membrane glycoprotein with a carboxy-terminal cytoplasmic domain, a transmembrane domain, and an extracellular region consisting of 2 immunoglobulin-like domains, namely, an amino-terminal immunoglobulin (Ig) variable-related domain (D1), which is distal to the cell surface, and a proximal IgC2 domain (D2)[1]. There is evidence for the evolutionary conservation of CAR, with homologues of the human receptor, hCAR, present in several mammalian species, including mice[2], rats, dogs, and pigs[3]. The presence of this protein significantly enhances the efficiency of adenoviral vector-mediated gene transfer[4]. However, the normal physiological function of this membrane protein is still not completely understood.
Recent studies suggest that CAR mediates homotypic intercellular adhesion as part of the tight and/or adherent junction[5]. CAR plays a role in tumorigenesis. In this respect, CAR expression has been correlated with both tumor suppression[6] and malignant transformation[7]. More recently, it has been shown that the expression of CAR in highly tumorigenic CAR-deficient human prostate cancer[8] and malignant glioma cells[6] leads to growth inhibition. Given that CAR is well positioned to participate in these roles, we investigated the effect of its overexpression on the human bladder cancer cell in vitro and in vivo.
Materials and methods
Cell culture and reagents Retroviral packaging cell line PT67[9], NIH3T3 fibroblasts, and human bladder cancer cell line T24 were obtained from the American Type Culture Collection (Manassas, VA, USA). The T24 cell line was established in 1970 from a urinary bladder carcinoma[10], which was identified as CAR-negative in the previous study[11]. The cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, NY, USA) at 37 °C supplemented with 10% fetal bovine serum (FBS). Retroviral vector pLXSN was purchased from Clontech Co (BD Biosciences Clontech, Palo Alto, CA, USA). The plasmid pTOPOCAR[4], including full length CAR cDNA, was obtained from Dr Jer-tsong HSIEH (Department of Urology, University of Texas Southwestern Medical Center, Dallas, TX, USA).
Construction of CAR-encoding retroviral vector An EcoR I/BamH I fragment encoding human CAR was obtained from pTOPOCAR and subcloned into a pLXSN backbone to generate the retroviral vector, named pLXSN-CAR.
Generation of viral particles and infection of target cells pLXSN and pLXSN-CAR were transfected into the PT67 packaging cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). PT67 packaging cells were split and grown in the selective medium with 500 mg/L G418. After 2 weeks, the G418-resistant colonies were picked and expanded in medium with 300 mg/L G418, and the supernatant viral particles were collected. The titer of viral stocks was determined by infection of the NIH3T3 cells[12]. The titer of pLXSN-CAR and pLXSN retrovirus was 1.6×109 colony-forming units/L and 2.0×109 colony-forming units/L, respectively.
For pLXSN-CAR retroviral transduction, the T24 cells were cultured and allowed to grow to subconfluence. The medium was then replaced with a 1:1 precipitated mixture of CAR retroviral supernatant and fresh complete medium. Polybrene (Sigma, St Louis, MO, USA) was added to the culture medium at a final concentration of 8 mg/L to enhance the binding of the virus to the host cells. The transduction was repeated the following day. Twenty-four hours post final transduction, the cells were harvested, resuspended, and maintained in selective medium containing 0.6 g/L G418 for up to 14 d. The G418-resistant cell clones were pooled, expanded, and tested for CAR expression. The control pLXSN retroviral transduction was performed as that of the pLXSN-CAR retrovirus. At last, 6 clones of CAR-positive cells and 8 clones of pLXSN cells were selected and expanded, respectively. Six CAR clones continuously expressed high levels of the CAR protein, so all the 6 CAR clones and 1 of the 8 pLXSN cell clones were applied for further study. The T24 cells successfully infected with the pLXSN-CAR retrovirus and the control pLXSN retrovirus were designated as T24/pLXSN-CAR and T24/pLXSN, respectively.
Western blotting and immunofluorescence staining Western blotting and immunofluorescence staining for the CAR protein in T24 cells infected with the CAR retrovirus and control virus were carried out as described in a previous report[13]. Briefly, a total protein extract (30 µg) isolated from harvested cells was then separated by 12% SDS-PAGE. Subsequently, the separated protein was transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA) and the membrane was blocked with 5% non-fat milk in TBS for 1.5 h. The membranes were then incubated for 1.5 h with the primary antibody against CAR at a dilution of 1:600 (Santa Cruz, CA, USA), and then hybridized for 1 h with the secondary antibody at a dilution of 1:5000. Immunodetection was performed using the ECL detection system (Pierce, Rockford, IL, USA). Protein loading equivalence was assessed by the expression of GAPDH.
To confirm the CAR expression, bladder cancer cells were fixed in 4% paraformaldehyde for 10 min and blocked with goat serum for 30 min, respectively. The cells were then incubated at 37 °C for 1 h with mouse anti-human CAR monoclonal antibody (Upstate Inc, Lake Placid, NY, USA) at a dilution of 1:150. After washing 3 times with PBS, the cells were incubated with tetramethylrhodamine isothio-cyanate (TRITC)-conjugated goat anti-mouse antibody at 37 °C for 1 h. The fluorescence staining intensity and intercellular location were then examined by fluorescence inverted microscope (Olympus IX 50, Tokyo, Japan).
Cell proliferation assay Cell growth was evaluated by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma, San Louis, MO, USA) assay. The bladder cancer cells (1×104/well) were plated in 96-well tissue culture plates in DMEM containing 10% FBS in a final volume of 0.2 mL. After incubation for 1, 2, 3, and 4 d, the cells were incubated with 20 µL MTT (at a final concentration of 0.5 mg/mL) at 37 ºC for 4 h. The medium was removed and the precipitated formazan was dissolved by adding 200 µL DMSO (Sigma, USA). After shaking for 10 min, the samples were lysed and the absorbance at 570 nm was detected using a Bio-Rad Technologies Microplate Reader (USA).
In vitro soft agar colony-formation assay T24/pLXSN-CAR1, T24/pLXSN-CAR2, T24/pLXSN, and parental T24 cells were collected and dispersed into a suspension of single cells in growth medium. One thousand cells were resuspended in 2 mL growth medium containing 0.3% low melting temperature agarose (Promega, Madison, WI, USA) and were overlaid in triplicate on 2 mL solidified 0.6% low melting temperature agarose in growth medium in a 6-well dish. The dishes were incubated for 2–3 weeks at 37 °C in a 5% CO2 atmosphere until colonies were formed. Colonies larger than 50 cells were then counted. Descriptive statistics (mean±SD) on colony sizes were calculated.
In vivo tumorigenicity assay T24/pLXSN-CAR1, T24/pLXSN, and parental T24 cells were harvested, washed, and resuspended in serum-free DMEM at the concentration of 5×107 cells/mL, and 1×106 cells were injected subcutaneously into 6–8-week-old BALB/c-nu/nu mice (n=5 mice per group, Shanghai Experimental Animal Center, Shanghai, China). Tumor dimensions were estimated based on bisecting diameters measured with a caliper, and the tumor volume was approximated using the formula 0.5 (ab2), where a is the long measured axis of the tumor, and b is the short measured axis of the tumor[14]. Tumor volume was measured at least weekly using the above formula. The mice were sacrificed after 5 weeks to measure tumor weight. All experiments on the animals complied with the Guidelines of Animal Care of Xi’an Jiaotong University.
Statistical analysis All data were expressed as mean±SD. Student’s t-test was used. P value <0.05 was considered statistically significant. All statistical tests were performed with statistical analysis software (SPSS, Chicago, IL, USA).
Results
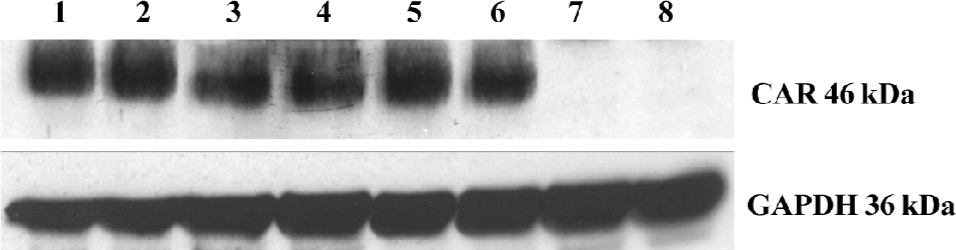
Determination of expression of CAR in the G418-resistant stable T24/pLXSN-CAR transfectant The pLXSN-CAR vector containing full-length CAR cDNA was successfully constructed. The expression of the CAR protein was detected by Western blot analysis and immunofluorescence staining. We found a high level of 46 kDa CAR expression in the T24/pLXSN-CAR cell clones. The CAR protein was undetectable in the parental T24 cells and control T24/pLXSN cells (Figure 1). Immunofluorescence staining was performed to confirm whether the retrovirus could infect and express the CAR protein in T24 cells. The intensity of the CAR-specific cell membrane immunofluorescence signal significantly increased in the T24/pLXSN-CAR cells, while in the T24/pLXSN and parental T24 cells, only a poor signal was observed (Figure 2).

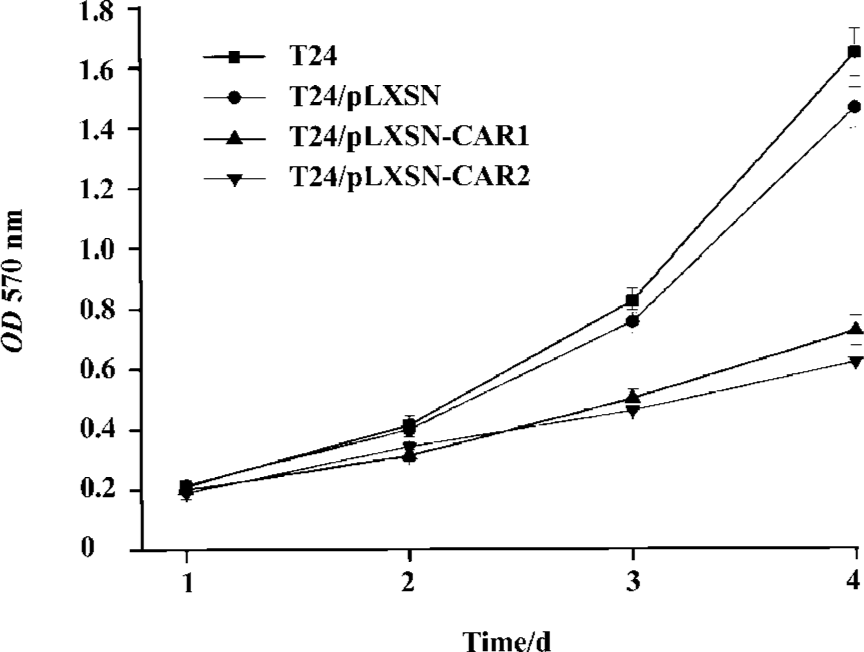
Effect of CAR overexpression on T24 cell growth To examine the effect of the CAR protein on the cell growth of T24 cells, we determined the growth rate of the CAR transfectant, control, and parental cells. Both T24/pLXSN and T24 cells exhibited a higher growth rate than T24/pLXSN-CAR1 and T24/pLXSN-CAR2, which was statistically significant (P<0.05; Figure 3B). Noticeably, no significant difference in the cell growth rate between the T24/pLXSN and T24 cells was detected (P>0.05).
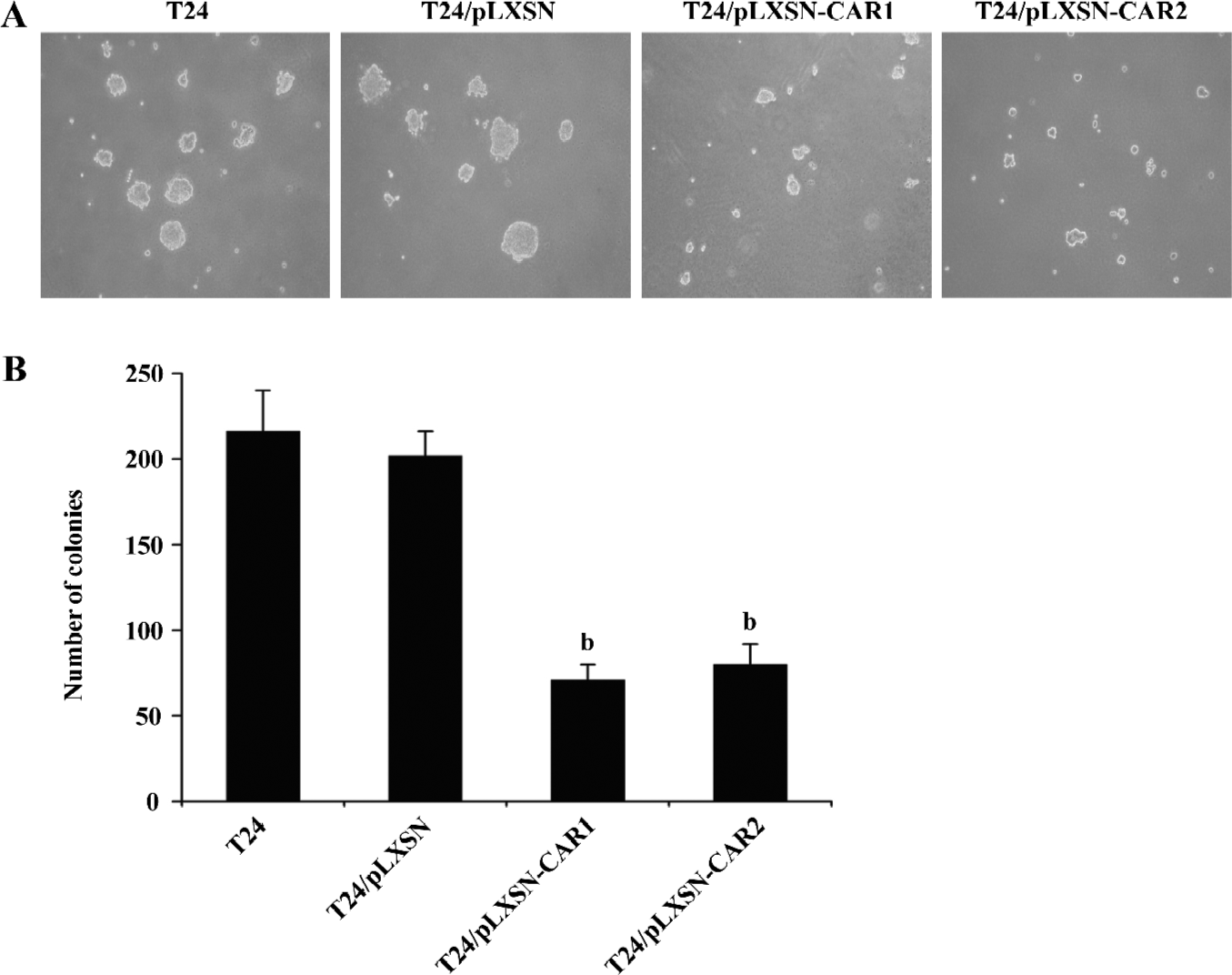
Effect of CAR on anchorage-independent growth The increased expression of CAR in T24 cells caused a significant reduction in the number of colonies formed by the T24/pLXSN-CAR1 cells (71±9) and T24/pLXSN-CAR2 cells (80±12) compared with the number of colonies formed by parental T24 or T24/pLXSN cells (216±24 and 202±14, respectively, P<0.05; Figure 4). In contrast, no significant difference was detected in the clone number between parental T24 cells and T24/pLXSN cells (P>0.05). Since anchorage-independent growth is considered in vitro tumorigenic assay, these data suggested that the overexpression of CAR could cause tumor inhibition in vitro.
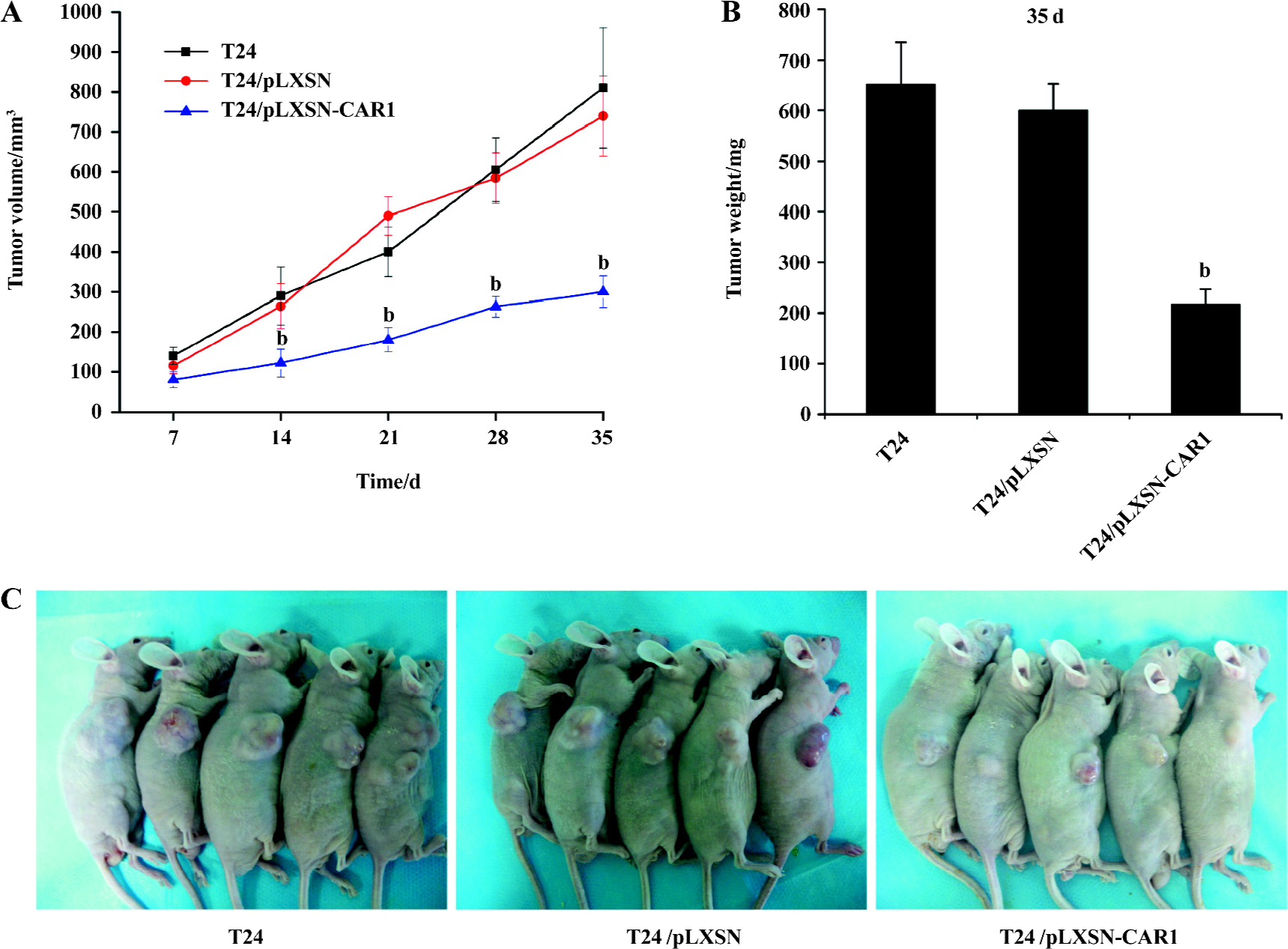
Reduced tumor growth of T24 cells with elevated CAR expression Tumor xenografts were established by subcutaneous injection of 1×106 cells from parental, control and CAR-transfected cells into the flank of 6–8-week-old BALB/c-nu/nu mice (n=5 mice per group). The mean tumor volume on d 7, 14, 21, 28, and 35 are shown in Figure 5A. T24/pLXSN-CAR1 tumors were much smaller than T24/pLXSN and parental T24 tumors (P<0.05). The volume of the T24 tumors was not significantly different from that of the T24/pLXSN tumors (P>0.05). On the thirty-fifth day after implantation, the average weight of the T24/pLXSN-CAR1 tumors was, significantly lower (217±30 mg) than that of tumors formed by the parental T24 cells (651±84 mg) and T24/pLXSN cells (600±52 mg, P<0.05; Figure 5B). The overexpression of CAR in T24 cells resulted in a significant inhibitory effect on the growth of the T24 tumors.
Discussion
CAR is a 46 kDa transmembrane glycoprotein, first identified as a high affinity receptor for both coxsackie and adenovirus (types 2 and 5)[1,2]. The presence of this protein significantly enhances the efficiency of adenoviral vector-mediated gene transfer[4]. Structurally, CAR is a typical Ig-like molecule with 2 Ig domains that interact with the adenovirus fiber protein. Most likely, the structure of this protein suggests that CAR may have other functions in addition to being a virus receptor[1]. Recent reports have indicated that CAR has homophilic interactions and appears to form a complex with ZO-1 in the tight junction of polarized epithelial cells[15]. Also, the loss of the CAR expression is associated with high grade cancer[16], and hence, it is involved in cell differentiation.
Using semiquantitative RT-PCR, an apparent down-regulation of CAR mRNA levels in the invasive tumors compared with superficial bladder cancer specimens has been documented[16]. Moreover, the study using immunohistochemical staining indicates that bladder cancers reveal a marked downregulation of CAR associated with stage and grade[17]. This suggests that altered CAR expression would be involved in the progression of bladder cancer. In addition to bladder cancer, the downregulation of CAR is also reported in ovarian cancer[18], melanoma[19], and head and neck cancer[20]. It has also been reported that the increased expression of the CAR gene leads to the inhibition of several of these tumors. However, the detailed characterization of the biological function of CAR is still lacking.
In the present study, we have examined the effect of CAR on bladder cancer cells based on cell growth in vitro and tumor growth in vivo. Using a highly efficient retroviral vector, we found that pLXSN-CAR-infected T24 cells grew much slower than the control retrovirus-infected or parental T24 cells. Noticeably, elevated CAR in T24 cells caused a significant reduction in the number of colony formations when T24 cells were plated on a semisolid agar plate. To support the in vitro results, animal xenografts have resulted in the same conclusion: tumors formed by T24 cells infected with the CAR retrovirus grow significantly slower than those by T24 cells infected with empty retroviral or parental T24 cells.
Our findings have demonstrated that CAR serves as a potent tumor suppressor in bladder cancer cells both in vitro and in vivo. Nevertheless, some reports suggest that the downregulation of CAR will reduce the volume of lung cancer in a mouse xenograft model[21], implying that the biological effect of CAR in different cancer types may vary. There-fore, further study is needed for elucidating the different mechanisms of the action of CAR in the development of different types of cancer.
Acknowledgement
We would like to thank Jer-tsong HSIEH (Department of Urology, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX, USA) for criticism and encouragement.
References
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997;275:1320-3.
- Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackie viruses. Proc Natl Acad Sci USA 1997;94:3352-6.
- Fechner H, Haack A, Wang H, Wang X, Eizema K, Pauschinger M, et al. Expression of coxsackie adenovirus receptor and alphav-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther 1999;6:1520-35.
- Li Y, Pong RC, Bergelson JM, Hall MC, Sagalowsky AI, Tseng CP, et al. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res 1999;59:325-30.
- Coyne CB, Bergelson JM. CAR: a virus receptor within the tight junction. Adv Drug Deliv Rev 2005;57:869-82.
- Kim M, Sumerel LA, Belousova N, Lyons GR, Carey DE, Krasnykh V, et al. The coxsackie and adenovirus receptor acts as a tumour suppressor in malignant glioma cells. Br J Cancer 2003;88:1411-6.
- Anders M, Hansen R, Ding RX, Rauen KA, Bissell MJ, Korn WM. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the coxsackievirus and adenovirus receptor. Proc Natl Acad Sci USA 2003;100:1943-8.
- Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res 2000;60:5031-6.
- Miller AD, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J Virol 1996;70:5564-71.
- Bubenik J, Baresova M, Viklicky V, Jakoubkova J, Sainerova H, Donner J. Established cell line of urinary bladder carcinoma (T24) containing tumour-specific antigen. Int J Cancer 1973;11:765-73.
- Lai YJ, Pong RC, McConnell JD, Hsieh JT. Surrogate marker for predicting the virus binding of urogenital cancer cells during adenovirus-based gene therapy. Biotechniques 2003;35:186-94.
- Bodine DM, McDonagh KT, Brandt SJ, Ney PA, Agricola B, Byrne E, et al. Development of a high-titer retrovirus producer cell line capable of gene transfer into rhesus monkey hematopoietic stem cells. Proc Natl Acad Sci USA 1990;87:3738-42.
- Li L, He DL. Transfection of promyelocytic leukemia in retro-virus vector inhibits growth of human bladder cancer cells. Acta Pharmacol Sin 2005;26:610-5.
- Guo QL, You QD, Wu ZQ, Yuan ST, Zhao L. General gambogic acids inhibited growth of human hepatoma SMMC-7721 cells in vitro and in nude mice. Acta Pharmacol Sin 2004;25:769-74.
- Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci USA 2001; 98: 15 191–6.
- Okegawa T, Pong RC, Li Y, Bergelson JM, Sagalowsky AI, Hsieh JT. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of car protein structure. Cancer Res 2001;61:6592-600.
- Sachs MD, Rauen KA, Ramamurthy M, Dodson JL, De Marzo AM, Putzi MJ, et al. Integrin alpha(v) and coxsackie adenovirus receptor expression in clinical bladder cancer. Urology 2002;60:531-6.
- Dmitriev I, Krasnykh V, Miller CR, Wang M, Kashentseva E, Mikheeva G, et al. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackie and adenovirus receptor-independent cell entry mechanism. J Virol 1998;72:9706-13.
- Hemmi S, Geertsen R, Mezzacasa A, Peter I, Dummer R. The presence of human coxsackievirus and adenovirus receptor is associated with efficient adenovirus-mediated transgene expression in human melanoma cell cultures. Hum Gene Ther 1998;9:2363-73.
- Li D, Duan L, Freimuth P, O’Malley BW Jr. Variability of adenovirus receptor density influences gene transfer efficiency and therapeutic response in head and neck cancer. Clin Cancer Res 1999;5:4175-81.
- Qin M, Escuadro B, Dohadwala M, Sharma S, Batra RK. A novel role for the coxsackie adenovirus receptor in mediating tumor formation by lung cancer cells. Cancer Res 2004;64:6377-80.
