Upregulation of Mark3 and Rpgrip1 mRNA expression by jujuboside A in mouse hippocampus1
Introduction
Jujuboside A (JuA) is a main effective component of jujubogenin, extracted from the seed of Ziziphus jujuba Mill var spinosa (Bunge) Hu ex H F Chou (Ziziphus), which is widely used in treating symptoms of insomnia and anxiety[1,2]. However, most previously published studies have only focused on the investigation of behavioral changes. Recent experimental results have suggested that a high dose of JuA could inhibit the hyperactivity of the hippocampal CA1 area induced by penicillin sodium[2]. JuA had inhibitory effects on the Glu-mediated excitatory signal pathway in the hippo-campus and hippocampal formation in vivo and in vitro[3,4]. Those studies suggested that JuA might play the role of an inhibitor on the central nervous system, especially on the hippocampus. However, they could not explain the functions of JuA on the hippocampus, and thus, the inhibitory effect of JuA on the hippocampus is still elusive.
In the present study, we adapted the protocol of the differential display polymerase chain reaction (DD-PCR) to screen for differentially-expressed genes modulated by JuA at the transcription level in the mouse hippocampus. We found genes modulated by JuA, and explained the partial molecular mechanism of JuA on the hippocampus at the gene transcription level. The use of DD-PCR is universal in investigating molecular pharmacological mechanisms of the effective component of a Chinese traditional herb at the gene transcription level.
Materials and methods
Chemicals and animals JuA was provided by the National Institute for the Control of Pharmaceutical and Biological Products in China (Beijing, China) with a purity above 98%. KM (Kunming strain) mice (male, Grade II, weighing 18–22 g) were obtained from the Laboratory Animals Center of Sichuan Academy of Medical Sciences (Chengdu, Sichuan, China), and maintained in an air-conditioned room with controlled temperature (23–25 °C) and humidity (50%–70%). The mice were housed in cages under a 12 h light/dark cycle with access to food and water. All the animal experiments were carried out from 19:00 PM to 23:00 PM. The animal experiments were performed with institutional ethical approval of protocol, and all efforts were made to minimize animal suffering. The mice were randomly divided between the control and the treated groups. The treated group was given an ip injection of JuA at 80 mg/kg, and the control group was given an equal volume of saline. Half an hour after the injection, the mice in the control (n=10) and treated groups (n=10) were observed for spontaneous activity. Meanwhile, the control group (n=6) and the treated group (n=6) were used for RNA extraction.
Spontaneous activity The mice were put in a ZZ-6 monitor (Chengdu Technology and Market Co, Ltd, Chengdu, Sichuan, China) for observation of spontaneous activity and adapted to the environment within 5 min. We then recorded the number of mouse movements within a 10 min period. After recording the number of mouse movement before any treatment, the mice were taken out and injected with JuA or saline and housed in the cages. Half an hour after the injection of JuA or saline, the frequency of mouse movement was recorded again using the same method.
RNA extraction All the animals were sacrificed by decapitation and the hippocampi were removed immediately. The total RNA of the hippocampus was isolated with Trizol (Molecular Research Center Inc, Cincinnati, OH, USA) according to the manufacturer’s protocol. To remove DNA contamination, the RNA samples were treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA) and extracted with phenol:chloroform. Finally, the RNA samples were resuspended in 50 µL of nuclease-free water. The concentrations of the total RNA were measured with a biophotometer, and the OD260/OD280 ratio of all RNA samples were up to 2.0.
DD-PCR The first strand cDNA was synthesized using the RevertAidTM First Strand cDNA Synthesis Kit (MBI Fermentas, Burlington, Ontario, Canada). For each reaction, 4 µg of deoxyribonuclease-treated total RNA was used for the reverse transcription, in a mixture of 2 µg 3'-anchored primers (T15A, T15C, and T15G, respectively) in a final volume of 12 µL, and heated at 70 °C for 5 min. Then 4 µL of 5× reverse transcription buffer, 1 µL of 20 U/µL, RibolockTM Ribonulease inhibitor (MBI Fermentas, Burlington, Ontario, Canada), and 2 µL of 10 mmol/L dNTP(dATP, dCTP, dGTP, dTTP) mix were added; the samples were incubated at 37 °C for 5 min. Then 1 µL (200 units) of RevertAidTM M-MuLV reverse transcriptase was added to each sample (with a final volume of 20 µL). The mixture was incubated at 42 °C for 60 min, and finally heated at 70 °C for 10 min.
PCR was performed in 25 µL reaction mixtures containing 2 µL cDNA, 2 µmol/L 3' anchored primers and 5' arbitary primers (Table 1), 200 µmol/L dNTP, and 2 units of Taq DNA polymerase, with an initial denaturation at 94 °C for 3 min, followed by 4 cycles at 94 °C for 1 min, 34 °C for 4 min, and 72 °C for 1.5 min, and 40 cycles at 94 °C for 1 min, 38 °C for 2 min, and 72 °C for 1.5 min, with a final extension at 72 °C for 10 min. After PCR amplification, 10 µL of PCR products were separated by electrophoresis on 8% denaturing polyacrylamide gels, and the gels were stained by silver[5].
Full table
Re-amplification, ligation, and transformation of PCR products and sequencing Differentially-expressed bands were retrieved from the polyacrylamide gels[6] and reamplified using the corresponding primer sets in 50 µL reaction mixtures containing 5 µL recycled DNA, and 1 µmol/L primers, respectively, 400 µmol/L dNTP and 5 units of Taq DNA polymerase, with an initial denaturation at 94 °C for 3 min, followed by 40 cycles at 94 °C for 1 min, 38 °C for 2 min, and 72 °C for 1.5 min, with a final extension at 72 °C for 10 min.
The reamplified PCR fragments were run on a 1% agarose gel and gel-purified using 3S Spin Agarose Gel DNA Purification Kit (Shenergy Biocolar, Shanghai, China) according to the kit protocol. Products were ligated into the pUCm-T cloning vector (Shenergy Biocolar, China) according to the kit protocol, then transfected into Escherichia coli high efficiency DH5α competent cells[7] with ampicillin and blue/white selection. The positive colonies were screened by PCR with a pair of M13 primers and digestion with restriction endonuclease Pst I. Those colonies were sequenced by a commercial sequencing service (Beijing Sunbiotech Co, Beijing, China) and the DNA sequence was analyzed with BLASTN (Bethesda, MD, USA; NCBI, National Center for Biotechnology Information).
Semi-quantitative RT-PCR The first strand cDNA was synthesized as protocol of DD-PCR except for 3'-anchored primers replaced by the oligo(dT)18 primer. PCR was performed in 25 µL reaction mixtures containing 1.5 µL cDNA, 2 µmol/L gene-specific forward and reverse primers (Table 2), 200 µmol/L dNTP, and 2 units of Taq DNA polymerase, with initial denaturation at 94 °C for 3 min, followed by 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min for the number of cycles optimized for each primer pair (Table 2) to ensure that the product intensity fell within the linear phase of amplification; final extension was at 72 °C for 10 min. RT-PCR amplification of β-actin transcript was used as the internal control to verify that equal amounts of RNA were used in each reaction. The PCR products were run on a 1% agarose gel, then photographed with a digital camera under UV illumination and analyzed by Quantity One software (Bio-Rad, Hercules, CA, USA).
Full table
Statistical analysis All data were expressed as mean±SD. A statistical analysis of the results was carried out by independent samples t-test and paired samples t-test. P<0.05 was considered significant.
Results
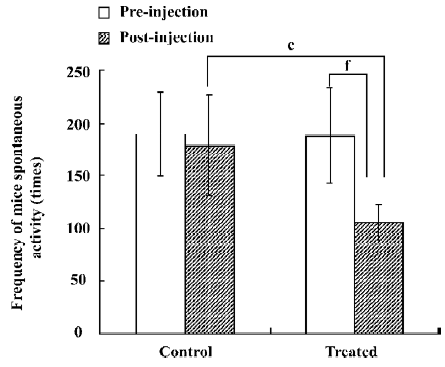
Analysis of spontaneous activity of mice The number of mouse movements in the control and treated groups before any treatment was 189.4±40.4 and 187.4±45.0, respectively. It was shown that there was no difference between the control and treated groups before the injection of saline and JuA (P>0.05). Half an hour after the injection of saline and JuA, the number of mouse movements in the control and treated groups was 178.9±47.5 and 105.7±16.8, respectively. The analysis of independent samples t-test between the control and treated groups after the injection of saline and JuA suggested that JuA significantly decreased the total activity intensity of the mice at a dosage of 80 mg/kg. The analysis of paired samples t-test between the preinjection and postinjection of JuA showed that JuA also decreased the total activity intensity of the mice significantly at the same dosage. Saline did not affect the number of mouse movements significantly (Figure 1).
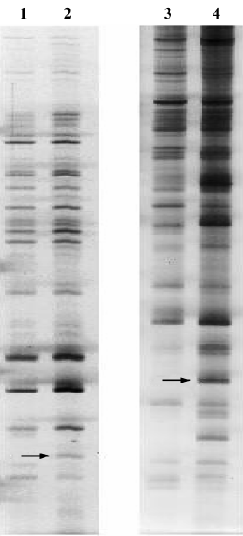
Analysis of differential expression in response to JuA by DD-PCR and confirmed by semi-quantitative RT-PCR To screen the differentially-expressed genes related to JuA in the mouse hippocampus, DD-PCR was performed (Figure 2). Following the isolation and sequencing of bands differentially-expressed between the control and treated groups and confirmed by semi-quantitative RT-PCR (Figure 3), we identified MAP/microtubule affinity-regulating kinase 3 (Mark3) and retinitis pigmentosa GTPase regulator interacting protein 1 (Rpgrip1) as upregulated genes by JuA in the mouse hippocampus.
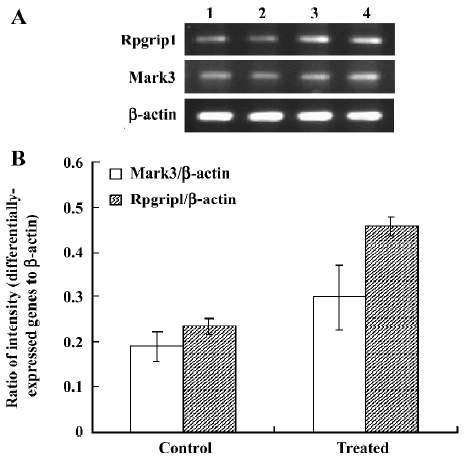
Discussion
We found that JuA, a component extracted from a Chinese traditional herb, significantly decreased total activity intensity in the mice. Our results were consistent with Feng et al[8], who confirmed that JuA had an inhibitory effect on the spontaneous activity of mice and increased quiet state time.
Why does JuA inhibit the spontaneous activity of mice? Although recent studies have suggested that JuA could inhibit the hyperactivity of the hippocampal CA1 area induced by penicillin sodium, with inhibitory effects on the Glu-mediated excitatory signal pathway in hippocampus[2,3], those studies could not explain why JuA had an inhibitory effect on the spontaneous activity of mice. In the present study, to elucidate the inhibitory effect of JuA on mice at the gene transcription level, we adapted the protocol of DD-PCR invented by Liang and Pardee[9] for detecting differentially-expressed genes in the mouse hippocampus, and the differentially-expressed genes were confirmed by semi-quantitative RT-PCR. Finally, we identified Mark3 and Rpgrip1 as upregulated genes by JuA in the mouse hippocampus.
Mark3 is also known as C-TAK1 (Cdc25C-associated kinase 1) or PAR1A[10,11], and has been implicated in cell cycle regulation and Ras signaling through its interactions with 2 putative substrates: the Cdc25C (cell division cycle 25C) phosphatase and the MAPK scaffold KSR1 (Kinase Suppressor of Ras 1)[10,12]. The upregulation of Mark3 mRNA by JuA may influence cell cycle or the Ras signaling pathway indirectly. Rpgripl, another differentially-expressed gene, is predominantly expressed and investigated in the retina[13–15]. It is also expressed in other regions including the heart, spleen, and brain[16], but has been poorly investigated in those regions. In the present study, we found that JuA could upregulate Rpgrip1 in the mouse hippocampus, which implies that Rpgrip1 plays an essential role in the hippo-campus.
In conclusion, JuA evidently had inhibitory effects on the spontaneous activity of mice; meanwhile, Mark3 and Rpgrip1 were upregulated by JuA in the mouse hippocampus. The relevance of JuA, the reduction of mice spontaneous activity, and the upregulation of Mark3 and Rpgrip1 exist in 2 possible pathways. One pathway is that JuA can upregulate Mark3 and Rpgrip1 directly or indirectly. The upregulation of Mark3 and Rpgrip1, as well as other reasons, can inhibit the spontaneous activity of mice. The other pathway is that JuA can inhibit the spontaneous activity of mice through an unknown way, and the inhibition on the spontaneous activity of mice could induce upregulation of Mark3 and Rpgrip1. However, although there are no studies to our knowledge which suggest the relevance between spontaneous activity and Mark3 or Rpgrip1, the two possible pathways suggest that there might be an essential link between the reduction of the spontaneous activity of mice and the upregulation of Mark3 and Rpgrip1.
References
- Hong G, Cao B. Advances in research on the seed of Zizyphus spinosa Hu. Zhong Yao Tong Bao 1987;12:51-3. Chinese..
- Shou CH, Wang J, Zheng XX, Guo DW. Inhibitory effect of jujuboside A on penicillin sodium induced hyperactivity in rat hippocampal CA1 area in vitro. Acta Pharmacol Sin 2001;22:986-90.
- Zhang M, Ning G, Shou C, Lu Y, Hong D, Zheng X. Inhibitory effect of jujuboside A on glutamate-mediated excitatory signal pathway in hippocampus. Planta Med 2003;69:692-5.
- Shou C, Feng Z, Wang J, Zheng X. The inhibitory effects of jujuboside A on rat hippocampus in vivo and in vitro. Planta Med 2002;68:799-803.
- Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 1991;196:80-3.
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Woodbury, NewYork: Cold Spring Harbor Laboratory Press; 2002.
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990;96:23-8.
- Feng ZY, Guo DW, Su S, Zhao H, Zheng XX. Sedative and anticonvulsant effect of jujuboside A. J Zhejiang Univ Medical Sci 2002;31:103-6. Chinese..
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 1992;257:967-71.
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ III, Wu Z, et al. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ 1998;9:197-208.
- Spicer J, Rayter S, Young N, Elliott R, Ashworth A, Smith D. Regulation of the Wnt signalling component PAR1A by the Peutz–Jeghers syndrome kinase LKB1. Oncogene 2003;22:4752-6.
- Müller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell 2001;8:983-93.
- Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet 2000;9:2095-105.
- Shu X, Fry AM, Tulloch B, Manson FD, Crabb JW, Khanna H, et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet 2005;14:1183-97.
- Zhao Y, Hong DH, Pawlyk B, Yue G, Adamian M, Grynberg M, et al. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci USA 2003;100:3965-70.
- Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet 2000;9:2095-105.
