Biotransformation of indomethacin by the fungus Cunninghamella blakesleeana1
Introduction
Indomethacin is a non-steroidal anti-inflammatory agent with anti-pyretic and analgesic properties. The anti-pyretic effect is caused by a resetting of the hypothalamic temperature-regulating center, whereas the anti-inflammatory and analgesic effects are due to inhibition of prostaglandin synthesis. It is extensively used because of its excellent pharmaceutical properties. Indomethacin is used in the treatment of musculoskeletal disorders such as rheumatoid arthritis, osteoarthritis, ankylosing spondylitis and also as an adjunct to periodontal therapy[1–3].
A few investigations on the metabolism of indomethacin in humans have been reported. In humans, indomethacin is metabolized by phase I metabolism to O-desmethylindo-methacin (DMI), N-deschlorobenzoylindomethacin (DBI) and O-desmethyl-N-deschlorobenzoylindomethacin (DMBI). These metabolites are devoid of anti-inflammatory activity. Indomethacin and all phase I metabolites can be conjugated by phase II metabolism into glucuronide conjugates[4–6].
An important factor in the evaluation of the efficacy and safety of a drug is the study of its mammalian metabolism. However, the identification of metabolites from animal sources and clinical samples may be hindered by insufficient quantities of material. Some micro-organisms can metabolize compounds in a similar manner to mammals, and have been used to isolate mammalian drug metabolites. With the low cost, ease of handling and scale-up capability, selected micro-organisms, particularly fungi, have been used successfully as in vitro models to mimic and predict the metabolic fate of xenobiotics in mammalian systems[7]. Using milder reaction conditions, the microbial system also provides an alternative to, or complement for, organic synthesis[8].
Although the metabolism of indomethacin in humans has previously been studied, to our knowledge, no report on the potential of filamentous fungi to metabolize indomethacin has been published. Cultures of five strains of Cunninghamella were chosen for this investigation, because such fungi, in previous studies in our laboratory, have the ability to metabolize drugs, including naproxen[9] and verapamil[10], in a similar manner to mammals. Therefore, the aim of the current study was to investigate the metabolic fate of indomethacin in cultures of five strains of Cunninghamella and to further compare the similarities between microbial transformation and mammalian metabolism.
Materials and methods
Chemicals Indomethacin (purity 99.5%) was purchased from Dongyu Pharmaceutical (Shenyang, China). Methanol and acetonitrile purchased from Concord Technology (Tianjin, China) were of HPLC grade. Peptone and yeast extract were obtained from Aoboxing (Beijing, China). All other chemicals were of analytical grade.
Micro-organisms and medium C elegans AS 3.156, C elegans AS 3.2028, C blakesleeana AS 3.153, C blakesleeana AS 3.910, and C echinulata AS 3.2004 were provided by the Institute of Microbiology, the Chinese Academy of Sciences (Beijing, China). Stock cultures were maintained on potato dextrose agar (Aoboxing, Beijing, China) slants at 4 °C and transferred every six months. First-stage fermentation was carried out in a medium consisting of 20 g of dextrose, 5 g of peptone, 5 g of yeast extract, 5 g of NaCl, 5 g of K2HPO4, and 1000 mL of distilled water. Second-stage fermentation was carried out in a wheat-bran medium with 1% wheat-bran in broth. The pH of two mediums were adjusted to 6.5 with 3.0 mol/L HCl, the mediums were then sterilized in Erlenmeyer flasks at 115 °C and 18 psi (pound per square inch) for 30 min, and cooled before incubation.
Fermentation procedures Microbial metabolism was facilitated by incubating the cultures with shaking on a rotary shaker at 28 °C. Fermentations followed a standard two-stage fermentation procedure. In general, for each of five strains of Cunninghamella, the first-stage of the fermentation was initiated by inoculating a 250-mL Erlenmeyer flask containing 50 mL broth with a loop of spores taken from a freshly growing agar slant. After incubation for 24 h, a 0.5 mL portion from the first-stage culture was used to inoculate a second-stage 100 mL flask containing 20 mL wheat-bran medium. The second-stage culture was incubated for 24 h, before the substrate in acetone-methanol (9:1, v:v) was added to a final concentration of 0.5 g/L. After 120 h of additional incubation, the cultures were centrifuged at 1500×g for 20 min, and the supernatant were decanted and kept at -20 °C until analysis.
Preparative-scale fermentation of indomethacin by C blakesleeana AS 3.910 followed the same procedure as the screening experiments except that the volume of broth was increased. During the first stage, two flasks were prepared as described above, and 100 second-stage flasks of 100 mL capacity containing 20 mL of wheat bran medium were each inoculated with 0.5 mL of the first-stage culture. Indomethacin was added to a final concentration of 0.5 g/L, and the culture was incubated for an additional 120 h.
In order to determine whether indomethacin would chemically decompose or spontaneously transform under fermentation conditions, substrate controls were prepared by adding the substrate to sterile medium and then incubating without the presence of micro-organisms. Culture controls consisted of a fermentation blank, in which the micro-organisms were grown under identical conditions without the addition of the substrate.
Extraction of metabolites and liquid chromatography-mass spectrometry (LC/MSn) assay A 0.1-mL aliquot of each sample was applied to a Bond Elute C18 cartridge (Teda Fuji, Tianjin, China), preconditioned by washing with 4.0 mL of methanol and 4.0 mL of distilled water. The cartridge was washed with 1.0 mL of distilled water, and metabolites were eluted with 1.0 mL of methanol. The eluate was evaporated to dryness under a gentle stream of nitrogen at 40 °C, and the residue was reconstituted by addition of 200 µL of acetonitrile-water (7:3, v:v). A 20-µL aliquot was injected onto the LC/MSn system.
LC/MSn analysis was performed on a Thermo Finnigan LCQ ion trap mass spectrometer (San Jose, CA, USA) equipped with an atmospheric-pressure ionization interface. The instrument was operated in positive electrospray ionization mode. The capillary voltage was fixed at 16 V, and the temperature was maintained at 200 °C. The spray voltage was set at 4.25 kV. The HPLC fluid was nebulized by using N2 as both the sheath gas at a flow rate of 0.75 L/min and the auxiliary gas at a flow rate of 0.15 L/min. A full-scan mass spectrum to obtain the protonated molecules [M+H]+ of each metabolite was collected. Multistage mass spectra were produced by collision-induced dissociation of the selected precursor ions with He present in the ion trap. The relative collision energy was set at 25%–35%. Data were collected and analyzed with the Xcalibur software (version 1.2; Thermo Finnigan). The metabolites were separated using a mobile phase consisting of acetonitrile-water-formic acid (70:30:0.5, v:v:v) over 20 min at 0.5 mL/min with a Diamonsil C18 column (200×4.0 mm inner diameter, 5 µm, Dikma) preceded by a Hypersil BDS-C18 precolumn (10×4.6 mm inner diameter, 5 μm, Dikma). The concentrations of indomethacin and its metabolites in cultures were determined by a validated LC/MS/MS method using glyburide as internal standard. The sample preparation, chromatography and mass spectrometry system were described above. Indomethacin and its metabolites were monitored using the following mass transitions: indomethacin, m/z 358→174 (collision energy 35 eV); M1, m/z 344→160 (collision energy 30 eV); M2, m/z 220→174 (collision energy 30 eV); M3, m/z 206→160 (collision energy 25 eV). Indomethacin and M1–M3 were quantified by their peak area ratio relative to that of the internal standard (glyburide, m/z 494→369; collision energy 35 eV) by weighted (w=1/c2) least-squares linear regression of calibration curves. The assay has been shown to be reproducible with precision below 15% and accuracy within ±15%. The lower limit of quantification was 50 ng/mL for the parent drug and all its metabolites.
Isolation and identification of metabolites The flask contents produced by the transformation of indomethacin using C blakesleeana AS 3.910 on a preparative scale were centrifuged at 1500×g for 20 min. The combined supernatant was extracted with three equal volumes of ethyl acetate. The organic extract was dried over sodium sulfate and evaporated to dryness under vacuum at 35 °C using a RE-52A rotary evaporator (Yarong Biology Instrument, Shanghai, China). In order to obtain sufficient quantities for the structural identification of metabolites, the methanol solution of extract was injected repeatedly onto a semi-preparative HPLC system (Shimadzu, Kyoto, Japan) consisting of two LC-6AD solvent delivery units, 2a DGU-14A degasser unit, an SCL-10A VP system controller, an SPD-10A VP UV-Vis detector, an FRC-10A fraction collector, and a CLASS-VP LC worksta-tion. Separation was accomplished using a mobile phase consisting of methanol-water (42:58, v:v) at 9.5 mL/min on a Shim-Pack PRC-ODS column (250×20 mm inner diameter, Shimadzu, Kyoto, Japan) preceded by a GPRC-ODS precolumn (8×1.5 mm inner diameter, Shimadzu, Kyoto, Japan). The UV detector was set at 230 nm. Compounds with similar retention times were pooled, evaporated to dryness, and stored at 4 °C before structural analysis.
Each metabolite was dissolved in 0.5 mL of dimethyl-d6-sulfoxide (99.96 atom% H) for nuclear magnetic resonance (NMR) analysis. The NMR measurements were carried out at 600 MHz on a Bruker ARX 600 NMR spectrometer (Bruker, Faellanden, Switzerland). Chemical shifts were recorded on the δ scales in parts per million (ppm) relative to tetrame-thylsilane as an internal standard.
Results
Screening of cultures Five strains of Cunninghamella were screened for the ability to catalyze the biotransformation of indomethacin. The results indicated that indomethacin was partially metabolized, and the percentages of transformation were C elegans AS 3.156 (38.76%), C elegans AS 3.2028 (69.29%), C blakesleeana AS 3.153 (51.90%), C blakes-leeana AS 3.910 (87.41%) and C echinulata AS 3.2004 (39.52%). Because the highest proportion of the indomethacin was transformed by C blakesleeana AS 3.910, this organism was selected for further investigation.
Identification of indomethacin metabolites The LC/MSn chromatograms of the culture control showed no metabolites or indomethacin present, and those of the substrate control showed only the presence of indomethacin but no metabolites. As shown in Figure 1, three metabolites and indomethacin were observed in the C blakesleeana AS 3.910 culture sample.
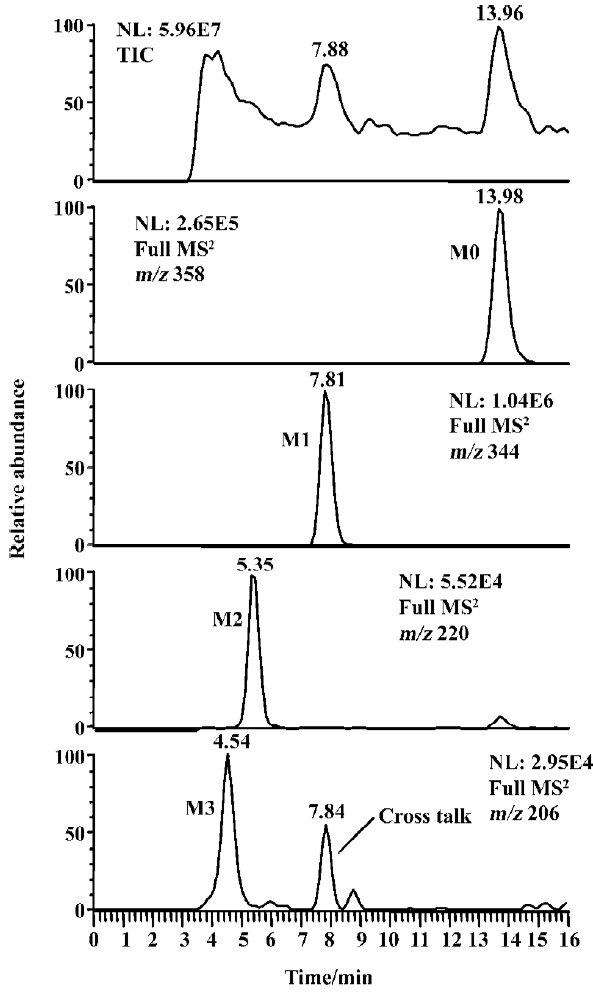
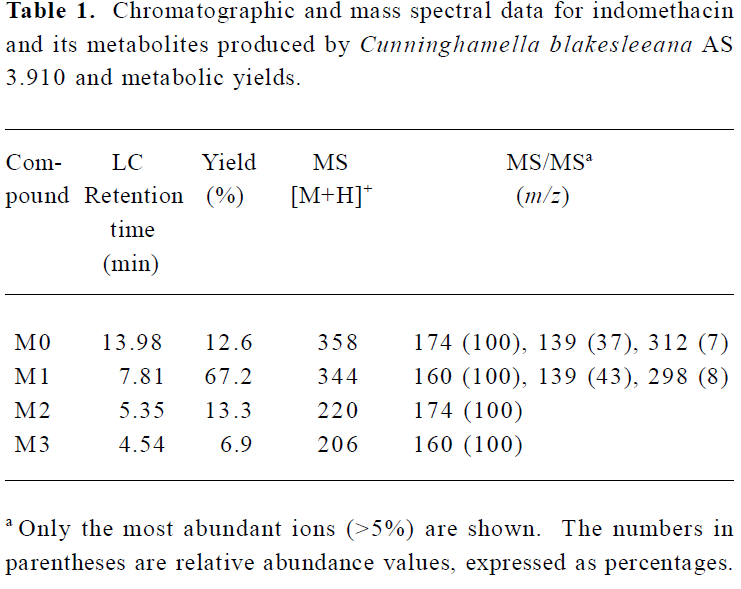
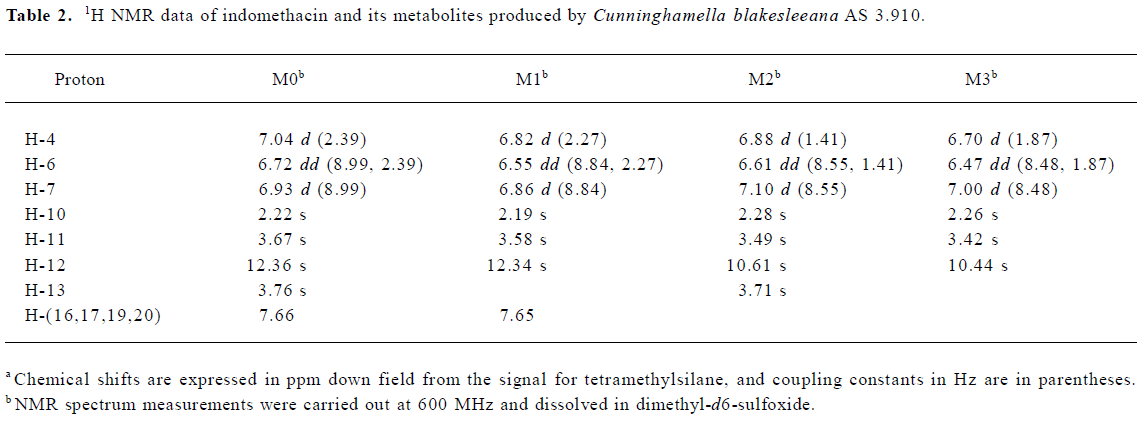
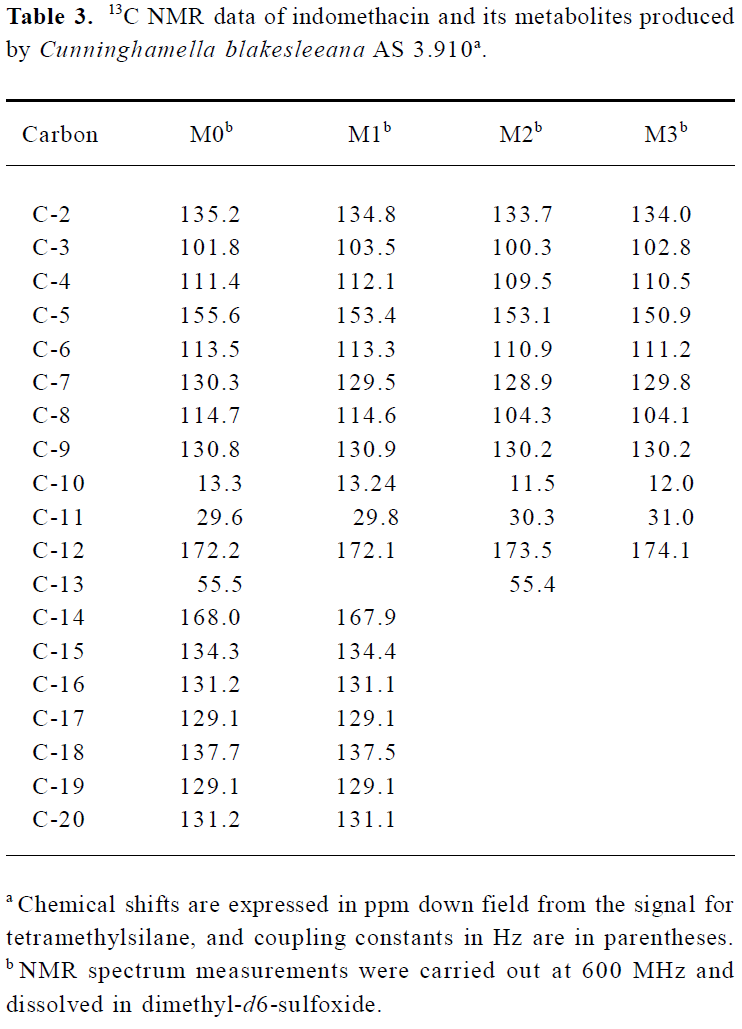
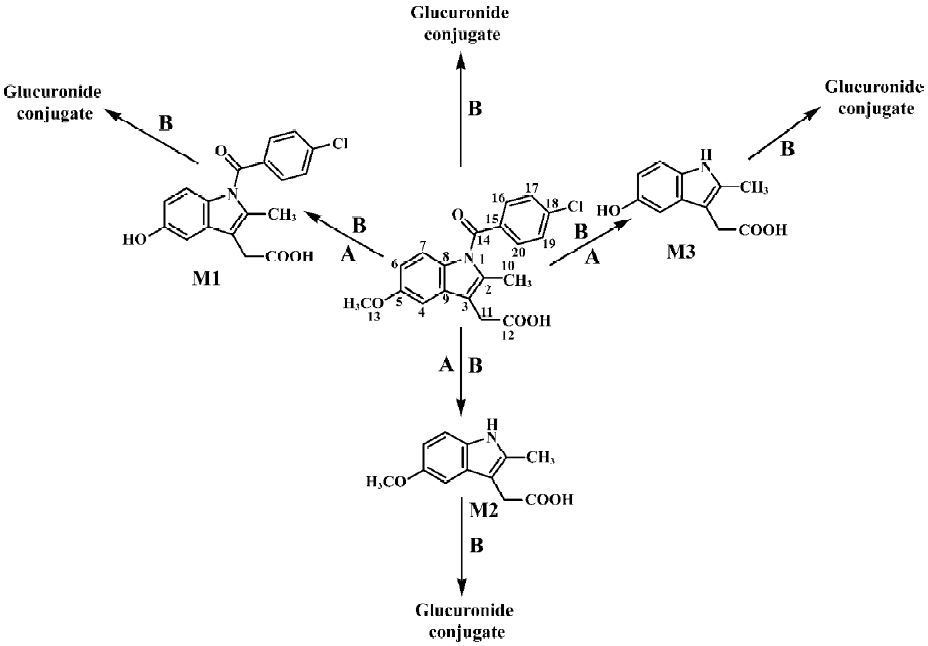
Following preparative-scale transformation of indo-methacin by C blakesleeana AS 3.910, three metabolites (M1–M3) were isolated by semi-preparative HPLC and their structures were identified by a combination analysis of LC/MSn (Table 1) and NMR (Table 2 and Table 3) spectra. The identity of M0 was confirmed by comparison of the LC reten-tion times and mass spectra (Table 1) with the synthetic references. The structures of indomethacin and its metabolites are shown in Figure 2.
Full table
Full table
Full table
Parent drug (M0) The compound eluting at 13.98 min by LC/MSn analysis were identified as indomethacin (M0), because the protonated molecules, full scan MS/MS spectra, and LC retention time of the compound were identical to those of the corresponding synthetic reference.
Metabolite M1 The retention time of M1 ([M+H]+ m/z 344) obtained by LC/MSn analysis was 7.81 min. The protonated molecule of M1 was 14 Da lower than that of M0. The diagnostic ions at m/z 160 for M1 from the parent ion at m/z 344 were 14 Da lower than the main fragment ion in the MS/MS spectrum of M0 (parent ion at m/z 358) (Table 1). This suggested that M1 was a monodesmethylated derivative. Comparisons of the NMR spectra of M0 and M1 indicated that M1 was O-desmethylindomethacin, based on the disappearance of the proton signal of 5-methoxy (Table 2) and the carbon signal of 5-methoxy (C-13, Table 3). Hence, M1 were deduced to be O-desmethylindomethacin.
Metabolite M2 The retention time of M2 ([M+H]+ m/z 220) obtained by LC/MSn analysis was 5.35 min. The protonated molecule of M2 ([M+H]+ m/z 220) was 138 Da lower than that of M0. The diagnostic ions at m/z 174 for M2 from the parent ion at m/z 220 were identical with the main fragment ion in the MS/MS spectrum of M0 (parent ion at m/z 358) (Table 1). This suggested that M2 was N-deschloro-benzoylindomethacin, which was further confirmed by NMR spectra of M2. Comparisons of the NMR spectra of M0 and M2 indicated that M2 was N-deschlorobenzoylindomethacin, based on the disappearance of the signal of the aromatic proton H-15 to H-20 (Table 2) and the disappearance of the signal of the aromatic carbon of chlorobenzoyl moiety (Table 3). Based on these MS and NMR data, metabolite M2 was identified as N-deschlorobenzoylindomethacin.
Metabolite M3 The retention time of M3 ([M+H]+ m/z 206) obtained by LC/MSn analysis was 4.54 min. The protonated molecule of M3 ([M+H]+ m/z 206) was 152 Da lower than that of M0. The MS/MS spectrum of M3 (parent ion at m/z 206) produced diagnostic ions at m/z 160, which was consistent with the main fragment ion in the MS/MS spectrum of M1 (parent ion at m/z 344) (Table 1). This suggested that M3 was O-desmethyl-N-deschlorobenzoylindomethacin, which was further confirmed by NMR spectra of M3. The disappearance of signal of aromatic protons in the chlorobenzoyl moiety and of the signal of 5-methoxy protons indicated that M3 was O-desmethyl-N-deschloro-benzoylindomethacin, which was also confirmed by the disappearance of signal of aromatic carbons in the chloroben-zoyl moiety and of the signal of 5-methoxy carbon. On the basis of these MS and NMR data, M3 was unambiguously identified as O-desmethyl-N-deschlorobenzoylindo-methacin.
Microbial transformation profiles and comparison with mammalian metabolism This study has shown that C blakesleeana transformed indomethacin to the following metabolites: O-desmethylindomethacin, N-deschloro-benzoylindomethacin and O-desmethyl-N-deschloro-benzoylindomethacin. In Figure 2, the structures of these metabolites and the proposed biotransformation pathways in micro-organism are compared to those identified in humans in previous studies[4–6]. After 120 h of incubation by C blakesleeana, approximately 87.4% of indomethacin was metabolized to three metabolites. The yields of those metabolites were M1 (67.2%), M2 (13.3%), and M3 (6.9%) (Table 1).
Discussion
We report here the successful biotransformation of indomethacin by C blakesleeana. As shown in Figure 2, After 120 h of incubation with C blakesleeana AS 3.910, approximately 87.4% of indomethacin was metabolized to three metabolites: O-desmethylindomethacin (DMI, M1, 67.2%), N-deschlorobenzoylindomethacin (DBI, M2, 13.3%) and O-desmethyl-N-deschlorobenzoylindomethacin (DMBI, M3, 6.9%). Three phase I metabolites of indomethacin produced by C blakesleeana AS 3.910 were identical to those obtained in humans.
It is well recognized that conjugations are important metabolic pathways of many compounds both in micro-organisms and in mammals. Conjugated metabolites (glucuronides, glucosides and sulfate conjugates) of drugs can be formed by microbial models[8–12]. Although indomethacin is excreted as glucuronides conjugates in humans[4–6], and previous studies using Cunninghamella species as models of mammalian drug metabolism indicated that these fungi were efficient at producing glucuronides[11] and sulfate conjugates[9] of drugs, we did not detect the glucuronide or sulfate conjugates of the above metabolites in the Cunninghamella species culture samples by LC/MSn analysis.
In conclusion, three metabolites of indomethacin were formed by C blakesleeana with the 67.2%, 13.3%, and 6.9% yields, respectively. The microbial transformation of indomethacin has some similarities with the metabolism of indomethacin in humans (Figure 2). Metabolites M1−M3 are phase I metabolites of indomethacin in both microbial models and in humans. In humans, O-desmethylindomethacin formation is critical to the elimination of indomethacin and represents 40%–55% of total drug eliminated in the urine[5]. As in humans, O-desmethylindomethacin was the major metabolite in the current microbial model. The microbial transformation of indomethacin further demonstrated the high potential of Cunninghamella to produce primary metabolites of a drug. The ability of Cunninghamella to mimic mammalian metabolism and to perform novel biotransformations clearly indicates that microbial systems represent an attractive alternative to the use of mammalian systems or chemical synthesis of metabolites. The quantities of drugs metabolites produced at low cost were sufficient for complete structural identification and for further use in investigating their pharmacokinetic, pharmacological and toxicological properties.
Acknowledgments
We thank Mr Ming-yu DUAN and Ms Li LI for their assistance on the conduct of the experiments.
References
- O’Brien WM. Indomethacin: a survey of clinical trials. Clin Pharmacol Ther 1968;9:94-107.
- Helleberg L. Clinical pharmacokinetics of indomethacin. Clin Pharmacokin 1981;6:245-58.
- Goodson JM, Dewhirst FE, Brunetti A. Prostaglandin E2 levels, and human periodontal disease. Prostaglandins 1974;6:81-5.
- Harman RE, Meisinger MAP, Davis GE, Kuehl FA Jr. The metabolism of indomethacin, a new anti-inflammatory drug. J Pharmacol Exp Ther 1964;143:215-20.
- Duggan DE, Hogans AF, Kwan KC, McMahon FG. The metabolism of indomethacin in man. J Pharmacol Exp Ther 1972;181:563-75.
- Vree TB, van den Biggelaar-Martea M, Verwey-van Wissen CP. Determination of indomethacin, its metabolites and their glucuronides in human plasma and urine by means of direct gradient high-performance liquid chromatographic analysis. Preliminary pharmacokinetics and effect of probenecid. J Chromatogr 1993;616:271-82.
- Srisilam K, Veeresham C. Biotransformation of drugs by microbial cultures for predicting mammalian drug metabolism. Biotechnol Adv 2003;21:3-39.
- Abourashed EA, Clark AM, Hufford CD. Microbial models of mammalian metabolism of xenobiotics: an updated review. Curr Med Chem 1999;6:359-64.
- Zhong DF, Sun L, Liu L, Huang HH. Microbial transformation of naproxen by Cunninghamella species. Acta Pharmacol Sin 2003;24:442-7.
- Sun L, Liu L, Huang HH, Zhong DF. Transformation of verapamil by Cunninghamella blakesleeana. Appl Environ Microbiol 2004;70:2722-7.
- Cerniglia CE, Freeman JP, Mitchum RK. Glucuronide and sulfate conjugation in the fungal metabolism of aromatic hydrocarbons. Appl Environ Microbiol 1982;43:1070-5.
- Chatterjee P, Pezzuto JM, Kouzi SA. Glucosidation of betulinic acid by Cunninghamella species. J Nat Prod 1999;62:761-3.
