Optimal design and validation of antiviral siRNA for targeting hepatitis B virus
Introduction
Hepatitis B virus (HBV) is the causative agent of acute and chronic hepatitis B in humans. To date, only about 20% of chronic HBV infection patients benefit from the conventional therapy (α-interferon and lamivudine) because of the low efficacy and drug-resistant mutants[1]. In recent years, a few new antiviral strategies have been developed as potential anti-HBV drugs[2,3].
RNA interference (RNAi) is now widely used to knockdown gene expression in a sequence-specific manner, making it a powerful tool not only for studying gene function, but also for therapeutic applications including antiviral treatments[4,5]. The replication of a wide range of viruses can be successfully inhibited using RNAi with both short interfering RNA (siRNA) and siRNA expression vectors[6,7]. However, for complicated virus genomes such as HBV, designing functional siRNAs that target viral sequences is problematic because of their extraordinarily high genetic diversity. It has been known that RNAi-resistant viral mutants emerge rapidly when targeting HIV sequences due to their high mutation rate[8–10]. Though no evidence has been reported about the RNAi-resistance for HBV, it is proposed that siRNA designed targeting conserved sequences among HBV eight genotypes could inhibit HBV virus replication effectively and is anticipated to resist potential viral mutational escape, since highly conserved sequences are likely to contain structurally or functionally constrained elements.
In addition, the optimal targets among the four open reading frames (ORF) of HBV in effectively inhibiting the HBV virus replication using RNAi were investigated in previous studies[11–16]. However, no studies have comprehensively compared the efficacy targeting different ORF of multiple siRNAs using quantitative structure-activity relationship (QSAR) analysis. The determination of the relationship between targeting ORF and efficacy of siRNAs could be helpful for optimal siRNA design targeting a complex overlapping genome, such as HBV.
The present study was designed to investigate the relationship between primary characteristics and siRNA efficacy by QSAR analysis, and to reveal the siRNA optimal design clues for complex overlapping genome HBV and further validated. We designed and evaluated quantitatively 23 siRNAs targeted throughout all four mRNAs, including the overlapping and non-overlapping regions on the HBV U95551 on both the S/P/Pregenomic mRNA and HBsAg level. Also, the degree of consensus within 8 genotypes of HBV of siRNAs was determined through plentiful computational analysis. Preliminary results showed that ORF S/P, X and genotype homology played important roles in inhibiting the S/P mRNA and HBsAg. However, C-target siRNAs contributed negatively to the expression of S/P mRNA and HBsAg. To clarify whether the different determined endpoints had correlation with the siRNA efficacy evaluation, the inhibition on C/P mRNA, HBeAg expressions and HBV DNA virus load by 23 siRNAs were evaluated. In summary, the contribution of the primary structure and different determined endpoints to the siRNA potency revealed in our study would be helpful for the siRNA optimal design for complex genome with overlapping ORF.
Materials and methods
Design and synthesis of siRNAs HepG2.2.15 human hepatoblastoma cell line was used as an experiment model[17]. The cell line was stably transfected with a head-to-tail dimer of HBV DNA genome (strain ayw, U95551) and could express all viral RNAs and proteins, viral genomes and secrete virus-like particles.
Twenty-three siRNAs were designed originally targeting U95551 throughout the HBV pregenomic RNA. The criteria used for siRNA design here involved: accordance with the basic design principle provided by online software; matching with the U95551 sequence; and, location in the relative conservative regions of part type B/C and U95551 sequences. The detailed process of the design was described as follows: the siRNAs involved in the study were selected from the siRNA pool offered by submitting U95551 sequence to several professional siRNA design websites (Ambion, Genepharm and Genesil) and in total, hundreds of selected siRNAs sequences were available. Then, on the basis of this designed siRNA pool, the homology of U95551 sequence and partial HBV genome sequences (type B and C) were analyzed. Finally, the 24 siRNAs were chosen from the relatively high conservative regions to avoid a high mutation part (supplementary materials 1).
All siRNAs were synthesized and purified by GenePharma Co (Shanghai, China), including the positive controls (1694 and 1826[18]; 2358 and 2422 [19]), and the negative control (NSC) siRNA. NSC siRNA had limited homology with any known sequences in the human, mouse and rat genomes.
Genotype homology of siRNA within HBV genome Primary nucleotide sequences of the 8 genotype HBV genomes were obtained from GenBank (URL:http://www.ncbi.nlm.nih.gov/htbin-post/Entrez) (15 typical sequences per genotype and total 120 sequences) (supplementary materials 2). Analysis of genotype homology, from genotype A to H, was determined by DNAMAN software (Lynnon Biosoft, Vaudreuil, Quebec, Canada) by aligning the siRNA and the 120 sequences. We defined 'conservation' as the percentage of sequence entries out of the 120 HBV sequences that showed perfect identity (ie, 21/21 matches) with the cognate 21-mer. Also, the genotype homology of each siRNA within HBV genotype from A to H was available.
Cell line and transfection HepG2.2.15 cells were maintained in MEM (Gibco-BRL, Grand Island, NY, USA) with 10% fetal bovine serum (FBS) (Hyclone, USA) and 200 µg/mL G418 (Gibco-BRL). Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. HepG2.2.15 cells were seeded in 48-well culture plates with 4×104 cells per well, and were transiently transfected with 10 nmol/L siRNAs. For mRNA and HBsAg examination, the cells and culture supernatants were harvested separately at 48 h. All assays were carried out 3 times, and values are expressed as mean±SD.
Real-time RT-PCR for determination of S/P mRNA expression Total RNA was extracted from HepG2.2.15 cells 48 h post-transfection using TRIzol reagent (TIANGEN, Beijing, China). RT-PCR was carried out using M-MLV reverse transcriptase (TAKARA), and 300–500 ng cDNA was used in SYBR Green real-time PCR (TIANGEN) for determining HBV gene expression in parallel with ribosomal protein LP0 (RPLP0) as an internal control. Primers for overlapping domains of the HBV S/P gene were 5′-ATCCTGCTGCTATGCCTCATCTT-3′ (forward) and 5′-ACAGTGGGGGAAAGCCCTACGAA-3′ (reverse), and for the housekeeping gene RPLP0 were 5′-TTAAACCCTGCGTGGCAATCC-3′ (forward) and 5′-CCACATTCCCCCGGATATGA-3′ (reverse). After a 10 min pre-incubation at 94 ºC, cDNA was amplified by 38 cycles of denaturation at 94 ºC for 30 s, annealing at 56 ºC for 30 s and extension at 72 ºC for 10 s. Melting curves were made to verify the amplification product specificity.
Primers for overlapping domains of the HBV C/P gene were 5′- CTCTGTATCGGGAAGC-3′ (forward) and 5′-TTAGGCCCATATTAGTG-3′ (reverse). The PCR protocol was the same except that the annealing temperature was 47 ºC.
The Pfaffl relative quantification method described previously[20] was used for determination of mRNA. All assays were repeated 3 times, and the final values were expressed as means±SD.
Quantitative analysis of HBsAg and HBeAg A quantitative enzyme-linked immunosorbent assay (ELISA) method using the sandwich enzyme immunoassay technique was developed to determine the HBsAg levels in cell culture supernatants. HBsAg quality control serum (10 ng/mL, National Center for Clinical Laboratory, NCCL) were used as a working standard to make standard curves using an HBsAg diagnostic kit (WanTai, Beijing, China), the absorbance measured was directly proportional to the concentration of HBsAg. Calibration curves of the ELISA assay appeared as a sigmoid curve, and the data were fitted to a 4-parameter logistic model by MicroCal Origin software (Origin Lab).
The methodology validation studies of HBsAg quantification revealed that the standard curves were stable with constant parameters. The inter- and intra-precision (CV%) of quality control (QC) samples were less than 20% and 10%, respectively. The specificity was appropriate for HepG2.2.15 (ayw). The dynamic range of the assay was 0.05–9.98 ng/mL, and the low limit of quantification (LLOQ) was 0.05 ng/mL, which was sufficient for the present study.
Similarly, the quantitative ELISA method was constructed by the HBeAg quality control serum (2 mg/mL) (NCCL) and HBeAg diagnostic kit (WanTai, Beijing, China). The methodology validation studies of HBeAg quantification showed that the inter- and intra-precision (CV%) of QC samples were less than 20% and 10%, respectively. The specificity was appropriate for HepG2.2.15 (ayw). The dynamic range of the assay was 0.2–16.6 ng/mL, and the low limit of quantification (LLOQ) was 0.2 ng/mL, which was sufficient for the present study.
Quantification of HBV DNA The 30 µL culture supernatant 48 h post transfection was harvested and boiled for 10 min to extract the total DNA. Real-time fluorescent quantification PCR was carried out to quantify HBV viral genomic DNA by using an HBV fluorescence quantitative PCR Diagnostic Kit (PG Biotech, China). The detailed procedure was carried out as the manufacturer’s protocol.
Results
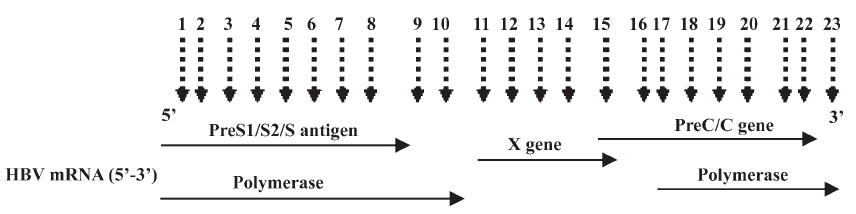
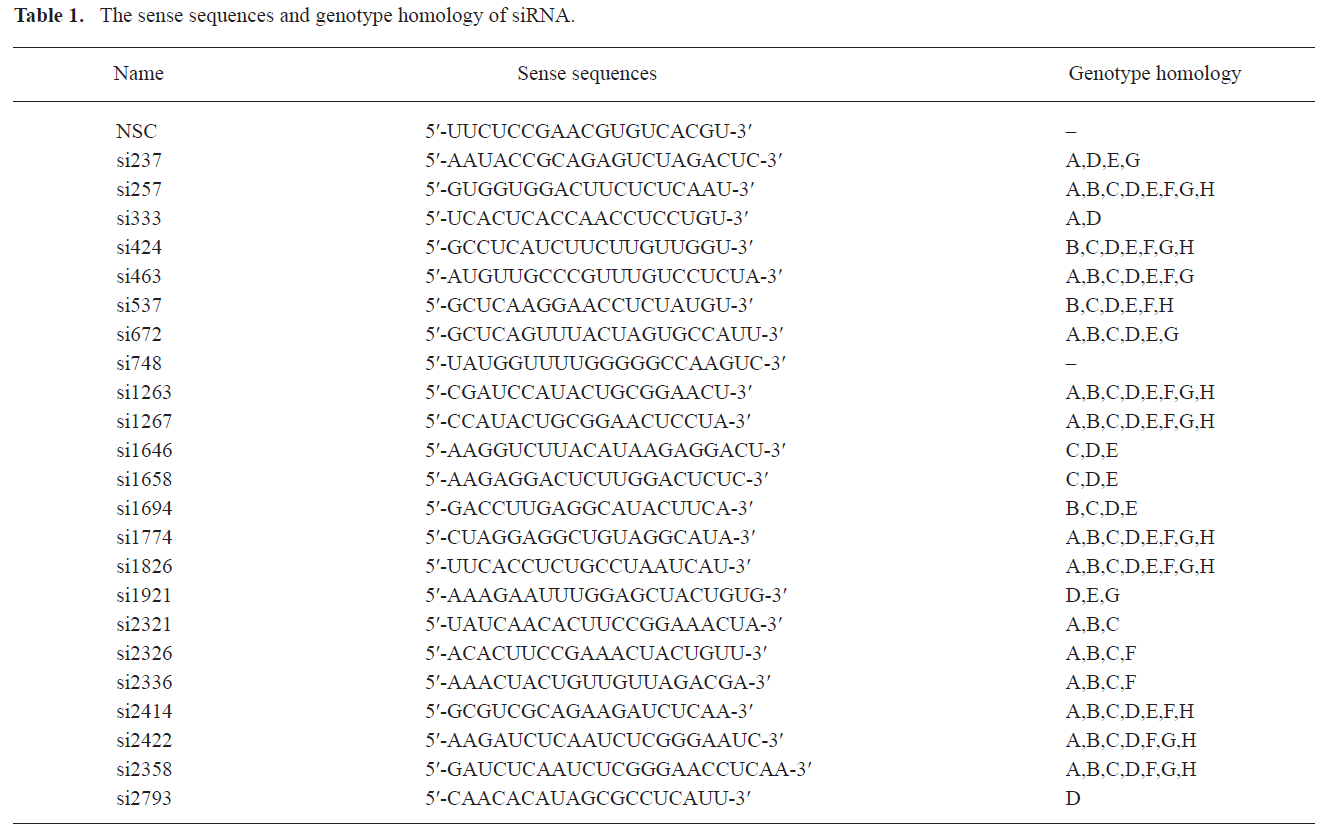
Design of multiple siRNAs and their characteristics Multiple sites targeting HBV 4 overlapping genes were designed (Figure 1). The oligo sequences coding for the sense strand of siRNA and the genotype homology are shown in Table 1.
Full table
Reduction of S/P mRNA and HBsAg expression by siRNA The 23 siRNAs exhibited diverse efficacy, with 17 out of 23 showing significant knockdown efficacy compared with the control group (P<0.05). The percentage of genotype homology of 23 siRNAs and their efficacy in inhibiting the S/P mRNA and HBsAg expression were analyzed (Figure 2).
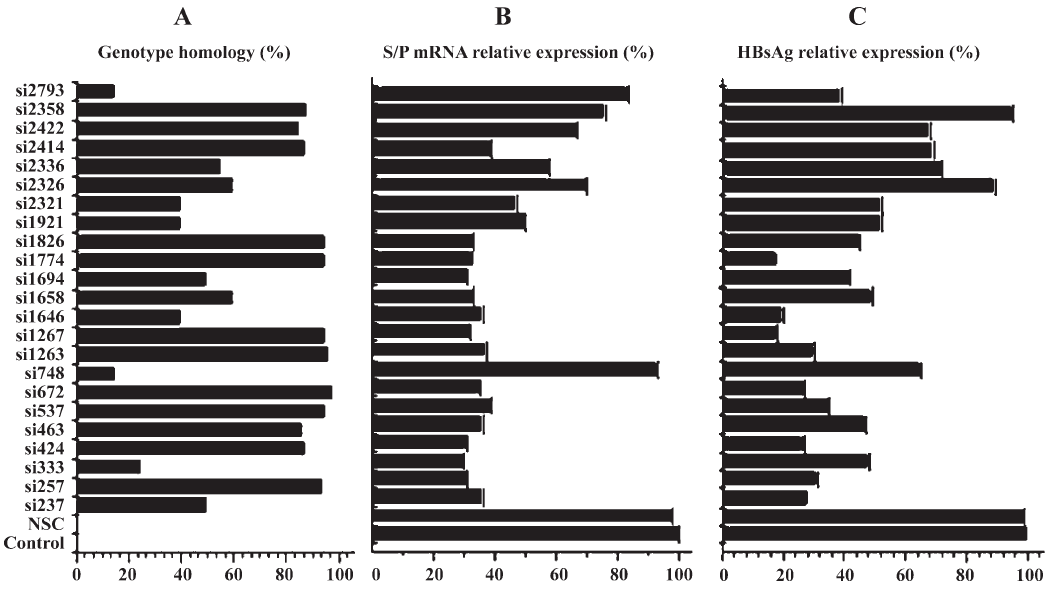
A multiple linear stepwise regression equation showed that the genotype homology, ORF S/P, X and C had tight correlation with the ability of siRNAs in inhibiting the expression of S/P mRNA and HBsAg, specially, C-target siRNAs played a negative role in inhibiting the two product expressions.
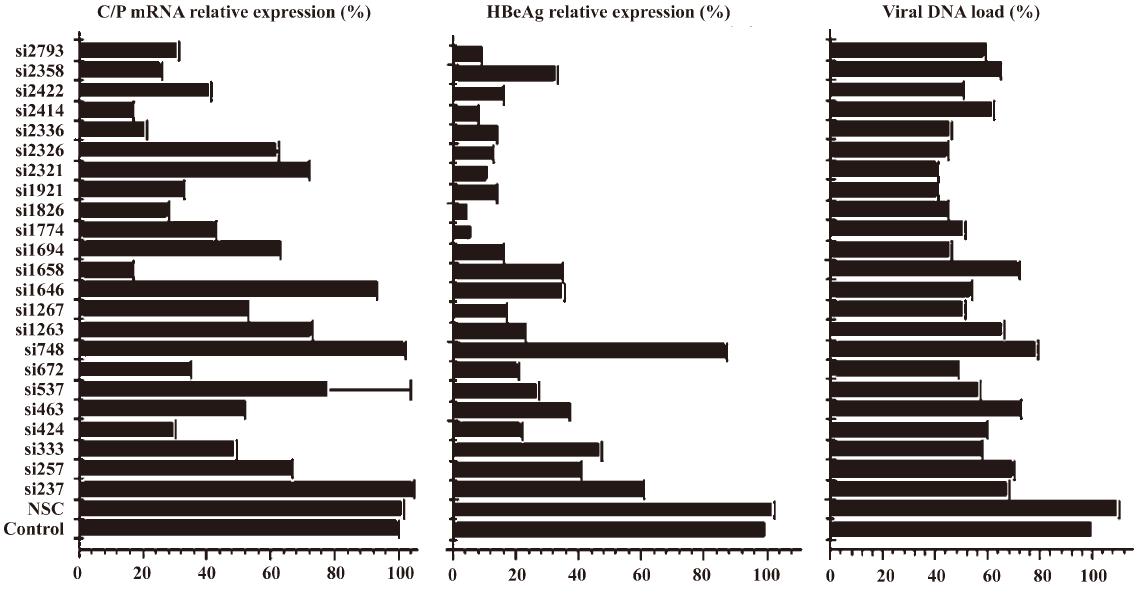
Relationship between determined endpoint and siRNA potency evaluation In the preliminary study, it was interesting that the siRNAs targeting ORF S/P instead of C were more effective in inhibiting the expression of S/P mRNA and HBsAg. To examine the relationship between different determined endpoint and siRNA efficacy evaluation, the ability of 23 siRNAs in inhibiting the expression of C/P mRNA, HBeAg, and the viral DNA load was investigated (Figure 3). Statistical results revealed that significant difference existed between siRNAs targeting ORF S and C in inhibiting different determined endpoints (C/P mRNA, HBeAg, S/P mRNA, HBsAg) (P<0.05). Generally, P-target siRNAs showed significant inhibition on the S mRNA and HBsAg expression. S-target siRNAs inhibited the expression of S mRNA and HBsAg strongly. X-target siRNAs played active roles in inhibiting all of the determined endpoints including S mRNA, HBsAg, C mRNA, HBeAg and viral DNA load. C-target siRNAs blocked the expression of C mRNA, HBeAg and viral DNA load significantly. Using ANOVA analysis, we statistically compared the silencing effect of any two siRNAs on mRNA, protein and HBV viral DNA expressions (Table 2).
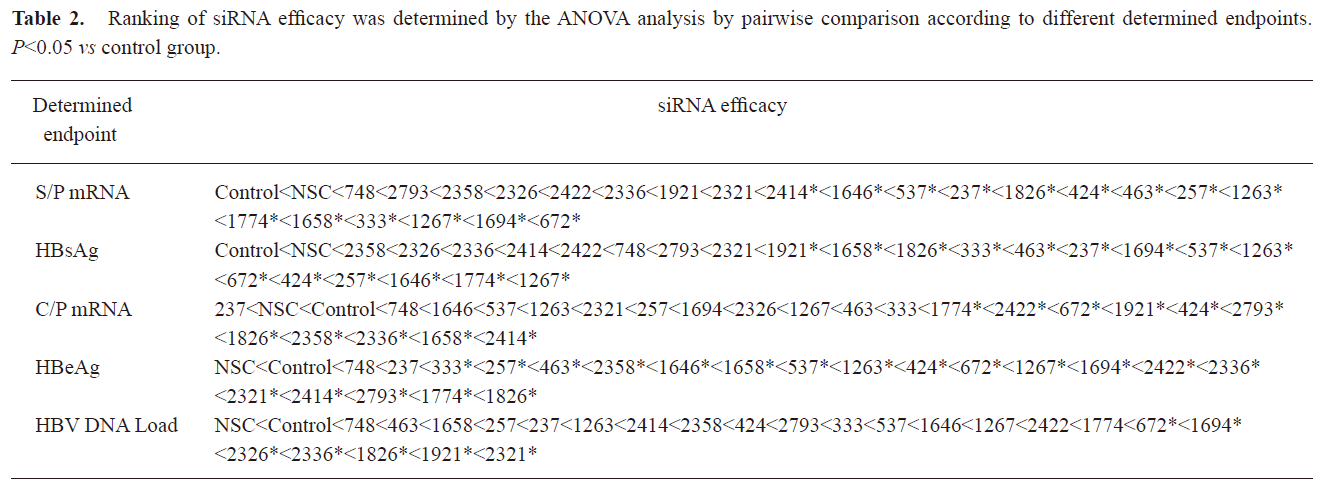
Full table
Discussion
In the present study, several premises, including synthetic siRNA, quantitative methods and ANOVA analysis were considered for selecting effective siRNA sequences as candidate sequences targeting HBV and QSAR analysis. Synthetic siRNA, rather than shRNA, was used, although shRNA is usually preferred by researchers because it is inexpensive and easy to use. However, the post-transcriptional gene silencing (PTGS) of synthetic siRNA is simple, more direct, and quantitative, because no in-vivo shRNA expression is involved in the transcription process. In the previous study, 10 shRNAs targeting different target sites of HBV pregenomic RNA were evaluated by semi-quantitative Northern methods[18]; however, no difference among the 10 shRNAs were available. In contrast, in our study successful ranking of si1694 and si1826 was made by the ANOVA analysis by pairwise comparison, which demonstrated the feasibility of the methods.
The correlation of genotype homology (or conservation) and optimal design of siRNA targeting HBV was validated in our study. A previous study had shown that RNAi-resistance could appear by mutation in RNAi-treated in HIV-1 infection[21]. For the complex virus genome with high mutations and multiple genotypes, such as HBV, HIV and HCV, optimal design of siRNA targeting conservative sequences was important not only for avoiding the potential RNAi-resistance but for the therapy of the multiple genotypes HBV infection. In another study[22], conserved regions within HIV-1 genomes were identified through an exhaustive computational analysis, and the functionality of siRNAs targeting the highest possible conserved regions was validated. The siRNA effectively inhibiting multiple genotypes of HIV-1 by targeting the best conserved regions in pandemic HIV-1 group M strains could be promising antiviral candidate for RNAi-resist HIV infection. Therefore, it is appealing that targeting genotype homology regions is important for siRNA optimal design.
In respect to candidate drugs screening, several siRNAs targeting the best conservative regions of HBV among genotypes from A to H showed satisfactory potency. Theoretically, these siRNAs with highly-genotype-homological targets, such as si257, si1263, si1267, si1774, and si1826, maybe inhibit the HBV infection despite genotype difference. However, HBV animal models with different genotype infection need to be constructed and whether the siRNA candidates targeting the conserved regions could inhibit every individual genotype of HBV also needs to be further investigated.
In our study, S/P mRNA and HBsAg were chosen as determined endpoints, and SPSS analysis showed that genotype homology had statistical correlations with the siRNA potency (P<0.05). Further SPSS analysis showed that genotype homology was also important for the inhibition of C/P mRNA, HBeAg and HBV DNA load expression, although the statistical significance did not reach the criteria (P=0.637, 0.884, 0.503 respectively). Two factors might contribute to this: the first might be the insufficient experimental repeats; the other might be due to the complex overlapping ORFs, which led to complicated results and disturbed the QSAR analysis.
The siRNA targeting different ORF could lead to different efficacy in inhibiting HBV expression and replication. Previous studies have shown that siRNAs targeting P, S, or X[12–16] ORF were effective, and we produced similar results in our structure-activity study. However, no studies comprehensively compared the inhibition diversity caused by siRNAs targeting different ORFs. In our study, statistical results showed that siRNAs targeting S/P and C/P ORF had relative stronger efficacy in inhibiting the corresponding expression product. One possible reason may be that siRNAs targeting different ORFs might have further influenced the different steps of HBV replication and thereby caused the differences in endpoint phenotypes determined. X-target siRNAs seemed important for the overall HBV replication progress, which may be related to the regulatory function of the X protein, and may also be related to the X gene that had highly-overlapping regions in the 3.5-, 2.4-, 2.1-, and -0.7 kb mRNA. Specially, the X-target and C-target siRNAs could significantly inhibit viral DNA load, which would be helpful for siRNA design for its drugable targeting HBV. Certainly, the phenomena need to be investigated further; however, the results in our study may be helpful for the siRNA optimal design for complex overlapping genes.
In summary, we proposed and validated a rational design of antiviral siRNA targeting highly divergent HBV. SiRNAs targeting different ORF could cause different determined endpoints on HBV expression and replication. Based on the current strategy in inhibiting HBV by RNAi, such as esiRNA and cocktails of siRNA, we expect that rational design of siRNA cocktails from ORF C, S, and X with high genotype conservation will be desirable to ensure greater efficacy and ensure against potential viral mutation. The preliminary results gave important hints to the optimal siRNA design for complex overlapping genes with high genetic diversity.
References
- Lau GK. Hepatitis B infection in China. Clin Liver Dis 2001;5:361-79.
- Tan C, Xuan B, Hong J, Dai Z, Hao R, Li Z, et al. RNA interference against hepatitis B virus with endoribonuclease-prepared siRNA despite of the target sequence variations. Virus Res 2007;126:172-8.
- Chen ZY, Cheng AC, Wang MS, Xu DW, Zeng W, Li Z. Antiviral effects of PNA in duck hepatitis B virus infection model. Acta Pharmacol Sin 2007;28:1652-8.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11.
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature 2004;431:371-8.
- Nielsen MH, Pedersen FS, Kjems J. Molecular strategies to inhibit HIV-1 replication. Retrovirology 2005;2:10.
- Leonard JN, Schaffer DV. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther 2006;13:532-40.
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol 2003;77:11531-5.
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol 2004;78:2601-5.
- Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an altern ative structure in its RNA genome. Nucleic Acids Res 2005;33:796-804.
- Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett 2003;543:51-4.
- Jiao J, Cao H, Chen XW, Zhou MJ, Liu ZH, Ding ZH. Downregulation of HBx mRNA in HepG2.2.15 cells by small interfering RNA. Eur J Gastroenterol Hepatol 2007;19:1114-8.
- Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther 2003;8:769-76.
- Liu SA, Wei HS, Dong QM, Guo JJ, Qin J, Zhang QY, et al. Inhibition of HBsAg and HBeAg secretion by RNA interference of the polymerase gene sequence of hepatitis B virus: an experimental study. Zhonghua Yi Xue Za Zhi. 2005;85:3079-83.
- Cheng TL, Chang WW, Su IJ, Lai MD, Huang W, Lei HY, et al. Therapeutic inhibition of hepatitis B virus surface antigen expression by RNA interference. Biochem Biophys Res Commun 2005;336:820-30.
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol 2003;21:639-44.
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA 1987;84:1005-9.
- Chen Y, Du D, Wu J, Chan CP, Tan YQ, Kung HF, et al. Inhibition of hepatitis B virus replication by stably expressed shRNA. Biochem Biophys Res Commun 2003;311:398-404.
- Li GQ, Gu HX, Li D, Xu WZ. Inhibition of Hepatitis B virus cccDNA replication by siRNA. Biochem Biophys Res Commun 2007;355:404-8.
- Lu PY, Xie F, Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet 2005;54:117-42.
- Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res 2005;33:2796-804.
- Naito Y, Nohtomi K, Onogi T, Uenishi R, Ui-Tei K, Saigo K, et al. Optimal design and validation of antiviral siRNA for targeting HIV-1. Retrovirology 2007;4:80.