Application of EGFP-EGF fusions to explore mechanism of endocytosis of epidermal growth factor1
Introduction
As a widely expressed housekeeping gene, the epidermal growth factor receptor (EGFR) plays an extremely important role in tumorigenesis[1]. During the last two decades, many studies have shown that the overexpression and mutation of EGFR have been frequently found in a variety of epithelial tumors, including non-small cell lung cancer, head and neck, breast, bladder, colorectal and ovarian cancers[2–5]. Several significant observations have established EGFR as a potential therapeutic target for the receptor-mediated gene delivery system in cancer treatment. In order to guide the receptor-mediated targeted gene therapy in clinical trials, we first need to explore the internalization mechanism of EGFR’s ligand. Epidermal growth factor (EGF) as an important ligand of EGFR is necessary to be labeled so that it is convenient to monitor protein localization of EGF in vitro and in vivo. The study of the internalization mechanism of EGF contributes to further understanding of the EGF-mediated gene delivery vectors endocytosis mechanism.
However, at present, EGF is usually labeled with radioactive reagents and fluorescein. These labeled EGF conjugates have some defects: the former has high radioactivity and a short half-life, and the latter is easy to quench and is unstable. To solve this issue, we made use of enhanced green fluorescent protein (EGFP) as an auto-fluorescent tag to label protein ligand EGF. EGFP is a unique GFP variant, which contains a chromophore mutation making the protein 35 times brighter than wild type GFP[6–8], so it greatly increases the sensitivity of the reporter protein than GFP[9,10]. Because of its inherent fluorescence, EGFP is ideal for experimental assays on living cells[11–13]. The detection of EGFP in living cells can be performed, and is amenable to a real-time analysis of molecular events[14–16]. The use of EGFP also eliminates the need for fixation or cell permeabilization and recombinant EGFP appears to retain its fluorescence and stability when produced as a fusion protein with foreign genes[17]. To explore whether EGF can be tagged with EGFP and remain its biological activity, we highly expressed the EGFP-EGF fusion protein in Escherichia coli (E coli). Cell-binding and internalization assay demonstrated that EGFP-EGF protein could bind to the cells expressing EGFR and be internalized. So this fusion protein was a very useful tool for investigating EGF internalization in vitro and in vivo.
Materials and methods
Reagents Restriction endonucleases were purchased from TaKaRa Biotechnology (TaKaRa, Dalian, China). Dulbecco’s modified Eagle’s medium (DMEM), Dulbecco’s phosphate-buffered saline (DPBS), fetal bovine serum (FBS) and trypsin/ethylenediamine tetraacetic acid were purchased from Invitrogen Corporation (Gibco, Grand Island, NY, USA).
Construction of pET28a-EGFP-EGF The EGFP gene was amplified from pEGFP-N1 using primers gfpNde: 5'-GGGAA-TTCCATATGGTGAGCAAGGGCGAG-3' and gfpEcor: 5'-CCGGAATTCCTTGTACAGCTCGTCCATG-3'. The PCR cycle conditions were 30 s at 95 °C, 30 s at 60 °C, and 50 s at 72 °C for 20 cycles, followed by a 5 min final extension step at 72 °C. The resulted DNA fragments were then digested with Nde I and EcoR I and ligated into pET28a Nde I/EcoR I-digested backbone. The resulted construct was called pET28a-EGFP. For the future insertion of other proteins into the carboxyl terminal of the EGFP, we then introduced the olignucleotides sequence (containing BamH I and Hind III restriction enzyme recognition site) MCSU: 5'-AATTCAGCC-AGCCGAAGCTTCGCGGATCCTAAC-3' and MCSD 5'-CGAGTTAGGATCCGCGAAGCTTCGGCTGGCTG-3' into the pET28a-EGFP EcoR I/Xho I backbone. The resulted vector was called pET28a-GFPmcs. The EGF gene was amplified from the M13KO7-EGF vector[18] by using primers EGF-2: 5'-CGCGGATCCGCGCAGTTCCCACCACTTC-3' and EGF-11: 5'-CCCAAGCTTAATAGTGACTCTGAATGTCCC-3'. The PCR cycle conditions were 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C for 20 cycles, followed by a 5 min final extension step at 72 °C. The resulted DNA fragments were then digested with BamH I/Hind III and then ligated into the pET28a-GFPmcs BamH I/Hind III backbone. The resulted vector was called pET28a-GFPmcsEGF. Thereafter, we introduced linker sequences (encoding GGGSGGGS) Link1: 5'-AGCTTAGA-GCCGCCGCCAGAGCCGCCGCCG-3' and Link2: 5'-AATTC-GGCGGCGGCTCTGGCGGCGGCTCTA-3' into the pET28a-GFPmcsEGF EcoR I/Hind III backbone, thus making vector pET28a-EGFP-EGF.
Protein expression and purification For the expression and purification of EGFP-EGF, the pET28a-EGFP-EGF plasmid was transformed into the E coli HMS174 (DE3) strain (Novagen, San Diego, CA, USA). A single colony was picked from a selective plate, inoculated into LB medium containing 100 µg/mL kanamycin and then incubated overnight at 37 °C with shaking. The cultures were diluted 1:20 into a selective LB medium, and then allowed to grow at 37 °C with vigorous shaking (~300 r/min), until the OD600 was 0.6. The cultures were then induced by adding IPTG (isopropyl-beta-D-thiogal-actopyranoside) to a final concentration of 1 mmol/L for 3 h at 30 °C, and the bacterial cells were harvested by centrifugation at 4000×g for 20 min at 4 °C. The bacterial pellet was then resuspended with 1×PBS and lysed by sonication on ice. To prevent protein degradation, the protease inhibitors (PMSF) were added at a final concentration of 1 mmol/L. The supernatant was collected after centrifugation at 12 000×g for 10 min at 4 °C. Then the cleared lysate was added 50% Ni-NTA (QIAGEN, Germany) slurry and mixed gently by shaking at 4 °C for 7–8 h. The lysate Ni-NTA mixture was loaded into a column and the column flow-through was saved for SDS-PAGE analysis. Then the protein was washed twice with wash buffer (20 mmol/L Tris-HCl, pH 7.9, 0.5 mol/L NaCl, 10% glycerol, 40 mmol/L imidazole) and the protein was eluted 4 times with elution buffer (20 mmol/L Tris-HCl pH 7.9, 0.5 mol/L NaCl, 10% glycerol) containing increasing concentrations of imidazole (80 mmol/L, 100 mmol/L, 200 mmol/L and 250 mmol/L). The wash fractions and the eluate were collected respectively for SDS-PAGE analysis.
Protein analysis and storage 12% SDS-PAGE was performed to identify the distribution of EGFP-EGF protein in every eluate. To further desalt and concentrate the EGFP-EGF fusion protein, the protein eluate were centrifuged at 4 000×g for 10 min at 4 °C with Amicon Ultra-4 centrifugal filter devices (Millipore Corporation, Billerica, MA, USA), and the concentrated solute were collected for SDS-PAGE analysis. The protein content was determined by bicincho-ninic acid protein assay (Pierce, Rockford, IL, USA). Finally, the purified protein was stored at -20 °C long term with HBS (20 mmol/L HEPES, 150 mmol/L NaCl, pH 7.4) buffer and 50% glycerol.
Western blot analysis Purified protein EGFP-EGF and EGFP were denatured and loaded on a 12% SDS-PAGE. Following separation, the protein was transferred to nitrocellulose filters (0.2 mm pore; Bio-Rad Laboratories, Hercules, CA, USA) in 192 mmol/L glycine, 25 mmol/L Tris base and 20% methanol. The filter was blocked in 5% non-fat milk in PBST for 2 h and incubated with rabbit anti-EGF polyclonal antibody (Santa Cruz Biotechnology, CA, USA) for 2 h and then with peroxidase-conjugated secondary antibodies (KangChen Bio-tech, Shanghai, China) for 1 h at room temperature. After washing with PBST, the filters were developed using a SuperSignal® West Pico kit (Pierce Biote-chnologies, Rockford, IL, USA) for 5 min, and exposed to X-ray film.
Cell culture HeLa cells (ATCC, Manassas, VA, USA) were cultured at 37 °C in DMEM supplemented with 10% FBS in a humidified atmosphere of 95% air and 5% CO2.
Binding assay of EGFP-EGF protein To examine the cell-binding activity of the EGFP-EGF protein, 3×104 HeLa cells were plated on 24-well dishes so that they were about 50% confluent the day of the experiment. The cells were then serum-starved for 3 h prior to the addition of the EGFP-EGF protein. EGFP-EGF was added to the cells at a final concentration of 200 ng/mL in DMEM. The cells were incubated at 4 °C for 1 h and then washed with PBS at room temperature. EGFP was used as a negative control. Competition assay was performed by co-incubating the cells with DMEM containing 200 ng/mL EGFP-EGF and 40 µg/mL EGF protein. Images were taken on fluorescence microscope (Axioskop 2; Carl Zeiss, Gottingen, Germany).
Internalization assay HeLa cells were maintained on dishes, detached, incubated with protein mixes and washed as outlined above in the cell-binding assay. After the final wash, the cells were placed at 37 °C to allow internalization activity. They were collected by centrifugation (1000×g) at the indicated time points after warming, resuspended in 0.1 mL of 2 mg/mL trypsin, 2 mmol/L EDTA and incubated at 37 °C for 1 min with agitation to remove cell surface-bound proteins. Trypsinized cells were centrifuged and washed 3 times with PBS, then fixed with 4% paraformaldehyde, resuspended in 0.5 mL PBS and analyzed by fluorescence-activated cell sorting (FACS).
Colocalization assay HeLa cells were seeded on cover-slips. The next day, the cells were serum-starved for 3 h and incubated with DMEM containing EGFP-EGF (the final concentration of EGF was 1.5 ng/mL) at 4 °C for 30 min, and then shifted to 37 °C for 2 h to allow internalization. The cells were then washed for 3×10 min with PBS, fixed and permeabilized with 4% paraformaldehyde, 0.1% Triton X-100 in PBS for 10 min, followed by washing with PBS again at room temperature. The cells were then blocked with 5% normal goat serum in PBS for 30 min, washed for 3×5 min with PBS, and then respectively incubated with mouse anti-clathrin mAb, mouse anti-early endosome antigen 1 (EEA1) mAb, mouse anti-lysosome-associated membrane protein 1 (LAMP1) mAb (Abcam, Cambridge, UK) and rabbit anti-EGFR polyclonal antibody (Santa Cruz, USA) diluted at 1:50 in PBS for 3 h at room temperature. After another wash of 3×10 min with PBS, tetramethylrhodamine isothiocyanate-labeled affinity purified goat anti-mouse IgG and goat anti-rabbit IgG (H+L; KPL, Guildford, UK) diluted at 1:50 in PBS was added and incubated for 30 min. Following a final rinse of 3×10 min with PBS, the cells were visualized under a Zeiss LSM confocal microscope.
Results
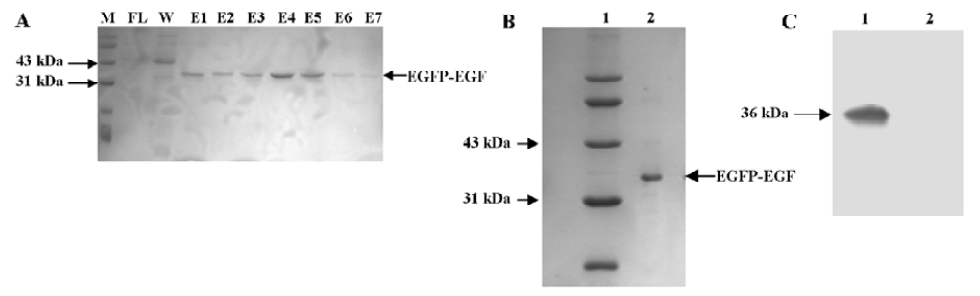
Expression and purification of the EGFP-EGF fusion protein The plasmid pET28a-EGFP-EGF containing 6×His (histidine) tag was constructed and the 36 kDa fusion protein EGFP-EGF was highly expressed in the E coli HMS174 (DE3) strain. Since EGFP is highly fluorescent even under normal light, we were able to visually follow the expression as well as purification of the protein. An SDS-PAGE analysis of the induced culture showed that the EGFP-EGF protein was expressed at very high levels, accounting for approximately 70%–80% of the soluble cytosolic proteins (data not shown). This could simplify the process of EGFP-EGF protein purification and maintain the biological activity of protein efficiently. Then this 36 kDa protein was purified by Ni-NTA resin and concentrated with Amicon Ultra-4 fentrifugal filter devices and finally analyzed by SDS-PAGE and Western blotting (Figure 1A–1C).
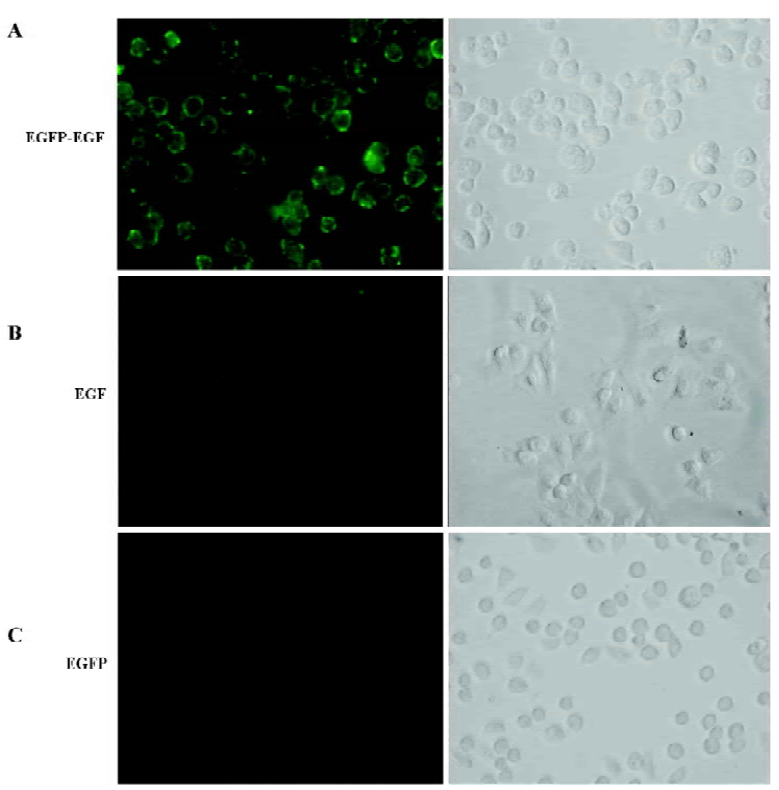
EGFP-EGF fusion protein retained receptor-binding activity The human cervix cancer cell line HeLa, which overexpresses EGF-containing receptors[20], was used to test receptor binding of EGFP-EGF. Serum-starved cells were incubated with EGFP-EGF at 4 °C for 1 h, then washed thoroughly to remove nonspecifically bound proteins and analyzed by fluorescence microscope. The result showed that EGFP-EGF protein could bind efficiently to the cell surface (Figure 2A). In order to determine whether the observed fluorescence is a result of specific ligand binding to cellular receptors and not caused by nonspecific binding to cell surfaces, we performed binding assay by co-incubating EGFP-EGF and excessive unlabeled EGF protein. We found that the binding activity of EGFP-EGF was significantly inhibited (Figure 2B). To further confirm receptor-binding specificity, we repeated the cell-binding assays using the EGFP protein and did not find cell binding (Figure 2C).
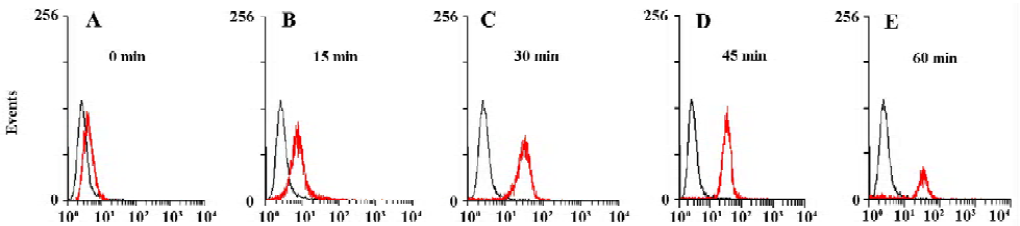
Establishing a FACS-based assay for internalization activity In order to determine whether the EGFP-EGF protein were internalized after binding, we measured intracellular accumulation of the EGFP-EGF protein. First, we allowed HeLa cells to bind EGFP-EGF proteins at 4 °C and washed the cells extensively to remove unbound proteins. The cells were then incubated at 37 °C and collected at sequential time points after warming. Each cell harvest was immediately followed by treatment with trypsin/EDTA to remove residual cell surface-bound proteins and then analyzed by FACS. The internalized EGFP-EGF protein was protected from trypsinization and so produced a fluorescence shift that was detectable by FACS. As shown in Figure 3A–3E, the HeLa cells treated with EGFP-EGF exhibited an increase in fluorescence shift over time, suggesting the accumulation of internalized EGFP-EGF.
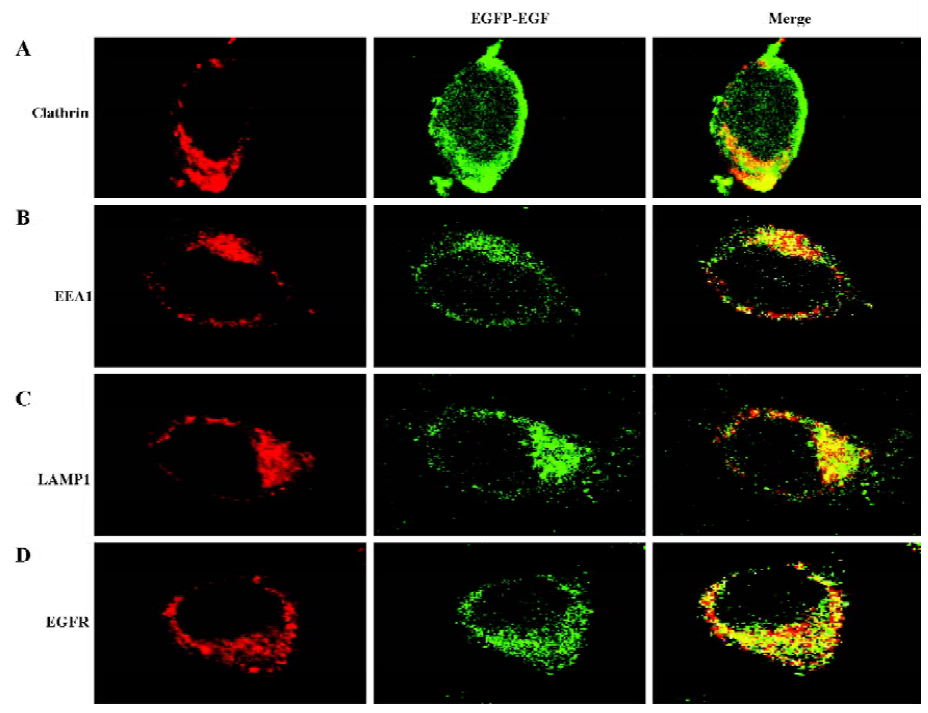
EGFR-EGF protein is efficiently internalized To further confirm that the EGFP-EGF protein can be efficiently internalized, we also examined the endocytosis pathway of EGFP-EGF by confocal microscope image. Previous studies have shown that low doses of EGF (1.5 ng/mL)[19,20] were predominantly internalized through clathrin-dependent pathways. In agreement with this finding, our results in Figure 4A clearly showed that EGFP-EGF (1.5 ng/mL EGF) was colocalized with clathrin. Furthermore, the overlap was observed between the internalized EGFP-EGF and early endosome marker EEA1 and late endosome marker LAMP1 (Figure 4B, 4C). Again, we stained the cells with an EGFR antibody for examining specific colocalization (Figure 4D). All these results demonstrated that the EGFP-EGF protein can be efficiently internalized into cells and has high sensitivity and biological activity.
Discussion
Because of EGFR’s great contribution in cell growth and development and relation with some severe diseases such as a wide range of epithelial derived cancers, it is often established as a potential gene therapeutic target, and the endocytosis mechanism of EGFR-mediated gene delivery vectors has been extensively studied. However, a lot of puzzles remain until now. One of them is the final fate of the internalization of EGF and EGF-mediated gene delivery vectors, so it is necessary to develop an available vehicle to facilitate the detection of EGF internalization in cells.
In this study, we successfully expressed and purified a 36 kDa EGFP-EGF protein which was identified by SDS-PAGE and Western blot analysis. To identify its biological activity and specificity, we performed a cell-binding and internalization assay in HeLa living cells. As shown in Figure 2, EGFP-EGF can efficiently bind to the cell surface and can be inhibited by the excessive unlabeled EGF protein. So the EGFP-EGF protein retains specific binding to receptor target and the EGFP-moiety does not interfere with the receptor-binding function. To confirm whether the EGFP-EGF protein can be efficiently internalized into cells, we also tracked the endocytosis pathway of the internalized EGFP-EGF through the early and late endosomes. As shown in Figure 4A–C, the overlap can be observed between the internalized EGFP-EGF and clathrin, early endosome protein EEA1 and lysosome-associated membrane protein LAMP1, which demonstrates that the EGFP-EGF protein can be actively endocytosed in living cells. Furthermore, the colocalization of EGFP-EGF and EGFR (Figure 4D) suggests that the EGFP-EGF receptor complex could be internalized simultaneously. These results suggest that this protein retained high EGFR binding bioactivity, efficacious targetability and specificity.
Our approach makes use of EGFP as an autofluorescent tag to label the EGF protein ligand. So we think that EGFP can be used to monitor cellular internalization of the EGF ligand under real-time conditions, and these activities can be measured using FACS. As shown in Figure 3A–E, we detected the internalization of EGFP-EGF by FACS. This assay approach predicts that a progressively increased accumulation of intracellular EGFP should produce a progressively enhanced fluorescence shift in the treated cell population over time. From these results, we found that most EGFP-EGF proteins were internalized by 30 min at 37 °C, suggesting rapid internalization after binding.
Taken together, we provide a more direct, convenient, reliable and feasible method in exploring the mechanism of EGF internalization. This fusion protein allows easy imaging and quantification using FACS or microscopy. This fusion protein has some advantages including high sensitivity, is hard to quench, easy preparation and high stability. It may enable researchers to visualize proteins within living cells without the need for chemical staining and avoiding light. All these traits make it a powerful tool to explore the mechanism of EGF endocytosis.
References
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 1995;19:183-232.
- Theodoropoulou M, Arzberger T, Gruebler Y, Jaffrain-Rea ML, Schlegel J, Schaaf L, et al. Expression of epidermal growth factor receptor in neoplastic pituitary cells: evidence for a role in corticotropinoma cells. J Endocrinol 2004;183:385-94.
- Sonnweber B, Dlaska M, Skvortsov S, Dirnhofer S, Schmid T, Hilbe W. High predictive value of epidermal growth factor receptor phosphorylation but not of EGFRvIII mutation in resected stage I non-small cell lung cancer (NSCLC). J Clin Pathol 2006;59:255-9.
- Xu YH, Richert N, Ito S, Merlino GT, Pastan I. Characterization of epidermal growth factor receptor gene expression in malignant and normal human cell lines. Proc Natl Acad Sci USA 1984;81:7308-12.
- Klijn JG, Berns PM, Schmitz PI, Foekens JA. The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review of 5232 patients. Endocr Rev 1992;13:3-17.
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996;173:33-8.
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature 1995;373:663-4.
- Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun 1996;227:707-11.
- Kimata Y, Iwaki M, Lim CR, Kohno K. A novel mutation which enhances the fluorescence of green fluorescent protein at high temperatures. Biochem Biophys Res Commun 1997;232:69-73.
- Cheng L, Fu J, Tsukamoto A, Hawley RG. Use of green fluorescent protein variants to monitor gene transfer and expression in mammalian cells. Nat Biotechnol 1996;14:606-9.
- Limon A, Briones J, Puig T, Carmona M, Fornas O, Cancelas JA, et al. High-titer retroviral vectors containing the enhanced green fluorescent protein gene for efficient expression in hematopoietic cells. Blood 1997;90:3316-21.
- Xia S, Kjaer S, Zheng K, Hu PS, Bai L, Jia JY, . Visualization of a functionally enhanced GFP-tagged galanin R2 receptor in PC12 cells: Constitutive and ligand-induced internalization. Proc Natl Acad Sci USA 2004; 101: 15 207–12.
- Campbell TN, Choy FY. Knockdown of chimeric glucocerebro-sidase by green fluorescent protein-directed small interfering RNA. Genet Mol Res 2004;3:282-7.
- He X, Li YL, Wang XR, Guo X, Niu Y. Mesenchymal stem cells transduced by PLEGFP-N1 retroviral vector maintain their biological features and differentiation. Chin Med J 2005;118:1728-34.
- Cinelli RA, Ferrari A, Pellegrini V, Tyagi M, Giacca M, Beltram F. The enhanced green fluorescent protein as a tool for the analysis of protein dynamics and localization: local fluorescence study at the single-molecule level. Photochem Photobiol 2000;71:771-6.
- Xia Z, Ye H, Locklin RM, Ferguson DJ, Cui Z, Triffitt JT. Efficient characterisation of human cell–bioceramic interactions in vitro and in vivo by using enhanced GFP-labelled mesenchymal stem cells. Biomaterials 2005;26:5790-800.
- Medina-Kauwe LK, Leung V, Wu L, Kedes L. Assessing the binding and endocytosis activity of cellular receptors using GFP-ligand fusions. Biotechniques 2000;29:602-9.
- Liang Y, Shi BZ, Zhang J, Jiang H, Xu YH, Li ZH, Gu JR. Better gene expression by (–)gene than by (+)gene in phage gene delivery systems. Biotechnol Prog 2006;22:626-30.
- Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem 2004;279:16657-61.
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA 2005;102:2760-5.
