Inhibition of Bcl-2 expression by a novel tumor-specific RNA interference system increases chemosensitivity to 5-fluorouracil in Hela cells
Introduction
RNA interference (RNAi) is a sequence-specific post-transcriptional gene-silencing process that has been proposed as a potential treatment for cancer[1]. Specific gene silencing can be achieved in a variety of cell systems using chemically synthesized siRNA or DNA vector-based shRNA[2,3]. A few promoters have been used to drive shRNA expression in cells, including the U6 and H1 RNA polymerase III promoters, and the cytomegalovirus (CMV) RNA polymerase II promoter.
However, the use of these promoters to drive shRNA expression in vivo would silence a given gene in all cell types, and thus produce undesirable effects in non-target cells. Cell-specific targeting of siRNA is an important issue to consider in RNAi therapy[4]. Before RNAi can be used to treat metastatic cancers, however, good cellular targets must be found[5]. To silence a gene specifically in tumor cells, without affecting its expression in normal cells, a tumor-specific promoter has to be used. To date, there are no reports of a tumor-specific promoter to drive shRNA expres-sion. Recently, cell-specific gene silencing has been achieved by using cell-specific promoters (SP-C) to drive shRNA expression, and a loxP-CRE regulatable RNAi system has also been developed[6,7].
In the present study, we attempted to develop a tumor-specific RNAi system directly under the control of the hTERT promoter. We investigated whether this system could depress the expression of exogenous reporter genes, firefly luciferase (LUC) and enhanced fluorescent protein (EGFP) and an endogenous gene (Bcl-2). We constructed corresponding vectors containing various shRNAs driven by hTERT promoters. Using these vectors, we demonstrated efficient silencing of target genes specific to tumor cells, but not in normal cell in vitro. We also established HeLa cells stably expressing shRNA to target Bcl-2, which could increase the chemosensitivity of cells to 5-fluorouracil.
Materials and methods
Plasmid construction To construct an hTERT promoter-driven shRNA vector (pBlock-TRTP-shRNA-mpA), the hTERT promoter, including 413 bp upstream from the transcription initiation site, was cloned from pGL3B-TRTP (a kind gift from Dr Mary CUSTER, National Cancer Institute, Bethesda, MD, USA) by polymerase chain reaction (PCR) and inserted into pSilencer4.1 (Ambion, USA) to replace the CMV promoter. A minimal poly(A) sequence (5'-TTATTTCCTAGAAAATAAAAGTAACCTAGACACACAACCAAAAAACATACGCCGGCGA-3') was used as a terminal sequ-ence. The DNA oligonucleotides coding for the shRNA were inserted between BamH1 and HindIII restriction sites. shRNAs targeting LUC, EGFP, Bcl-2, and an unrelated shRNA negative control (control) were synthesized: shLUC, sense 5'-GATCCCTTACGCTGAGTACTTCGATT CAAGA-GATCGAAGTACTCAGCGTAAGTTA-3', antisense 5'-AGCTTAACTTACGCTGAGTACTTCGATCTCTTGAATCGAA-TACTCAGCGTAAG-3'; shEGFP, sense 5'-GATCCCACAA-GCTGGAGTACAACTACTTCAAGAGAGTAGTTGTACTCCA- GCTTGTGTTA-3', antisense 5'-AGCTTAACACAAGCT-GGAGTACAACTACTCTCTTGAAGTAGTTGTACTCCA-GCTTGTGG-3'; shBcl-2, sense 5'-GATCCGTACATCCATTA-TAAGCTGTTTCAAGAGAACAGCTTATAATGGATGTA-CTT-3', antisense 5'-AGCTTAAGTACATCCATTATAA-GCTGTTCTCTTGA AACAGCTTATAATGGATGTACG-3'; shControl, sense 5'-ATCCACTACCGTTGTTATAGGTG-TTCAAGAGAC ACCTATAACAACGGTAGTA-3', antisense 5'-AGCTTACTACCGTTGTTATAGGTGTCTCTTGAACA-CCTATAACAACGGTAGTG-3'. The target sequences were selected based on published synthetic siRNA sequences[6,8,9]. To facilitate the analysis of shRNA efficacy, an sv40-driven luciferase-expressing cassette from pGL3-promoter was separately inserted into pBlock-TRTP-shLUC-mpA and pBlock-TRTP-shControl-mpA between SalI and NotI restriction sites. The CMV-driven EGFP-expressing cassette from pGL3-CMV-EGFP (constructed with a framework of pGL3-promoter, with the CMV promoter from pCDNA3.0 replacing the sv40 promoter between BglII and HindIII restriction sites, and the EGFP gene from pEFGP-N1 replacing the luciferase gene between HindIII and XbaI restriction sites) was separately inserted into pBlock-TRTP-shEGFP-mpA and pBlock-TRTP-shControl-mpA between SalI and NotI restriction sites. The method used was similar to the one described by Zeng et al[10].
Cell culture and transfection HeLa, HepG2, and A549 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum (BCS), 100 U/mL penicillin and 100 mg/mL streptomycin. WI-38 cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin. The cells were transfected in opti-MEM with the transfection reagent Lipofectamine-2000 (Invitrogen, USA) according to the manufacturer’s instructions. To establish HeLa cells stably expressing shRNA, cells were treated with 2 µg/mL puromycin (Sigma, USA) for 14 d, and after that cells continued to be cultured with 1 µg/mL puro-mycin.
Luciferase assay HeLa, HepG2, A549 and WI-38 cells were transfected in 24-well culture plates with 0.8 μg firefly luciferase (Fr-luc)-expressing vectors (pBlock-TRTP-shLUC-mpA-sv40-luciferase or pBlock-TRTP-shControl-mpA-sv40-luciferase) and 0.01 μg Renilla luciferase (Re-luc)-expressing vector (pRL-sv40) as transfection controls. The cells were harvested 48 h after transfection. The luciferase activity was measured with a dual luciferase assay kit (Promega, USA) using a luminometer (Lumat LB 9506, Berthold Technology). The relative luciferase activity was defined as the ratio of Fr-luc activity to Re-luc activity.
Reverse transcription polymerase chain reaction analysis After being treated with DNase, 2 µg of total RNA was reverse-transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen, USA) in the presence of random primers. The cDNA was used to amplify the EGFP or Bcl-2 fragments. For normalization of RNA loading, the housekeeping gene β-actin was also amplified from each sample. The primer sequences were as follows: EGFP, sense 5'-TGCCACCTACGGCAAGCTGA-3', antisense 5'-TCGA-TGTTGTGGCGGATCTT-3'; Bcl-2, sense 5'-CGACGA-CTTCTCCCGCCGCTACCGC-3', antisense 5'-CCGCATGC-TGGGGCCGTACAGTTCC-3'; β-actin, sense 5'-CCAAGG CCAACCGCGAGAAGATGAC-3', antisense 5'-AGGGTA-CATGGTGGTGCCGCCAGAC-3'. PCR amplification was performed using the following conditions: 1 cycle of 95 °C for 5 min; for EGFP 25 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s; for Bcl-2, 30 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s; followed by a final elongation step of 72 °C for 10 min. After amplification, 10 µL of PCR product was run on a 1.5% agarose gel and visualized by ethidium bromide staining.
Fluorescence-activated cell sorting analysis HepG2, HeLa, A549, and WI-38 cells were transfected in 6-well culture plates with pBlock-TRTP-shEGFP-mpA-CMV-EGFP or pBlock-TRTP-shControl-mpA-CMV-EGFP. Forty-eight hours after transfection, the cells were photographed by using a fluorescence microscope and then harvested. Half of the cells were used to extract total RNA for reverse transcription (RT)-PCR, the others to observe the levels of EGFP fluorescence using fluorescence-activated cell sorting (FACS; calibur, BD, USA).
The cell cycle distribution profile was analyzed by FACS. A total of 1×106 cells were collected and washed with phosphate-buffered saline (PBS), fixed in ice-cold 70% ethanol and stored at 4 °C. After being resuspended in PBS containing 100 μg/mL RNase A and 20 μg/mL propidium iodide (PI) for 30 min, samples were analyzed by FACS.
MTT assay The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was performed to assess the effect of Bcl-2 on cell proliferation and chemosensitivity to 5-fluorouracil. Cells were plated in 96-well plates at a density of 5×103 cells per well. Then for 7 d, every 24 h a batch of cells were stained with 20 µL sterile MTT dye (5 mg/mL; Sigma, USA) at 37 °C for 4 h, then culture medium was removed and 150 µL of Me2SO was added and thoroughly mixed in for 10 min. Spectrometric absorbance at 570 nm was measured by using a microplate reader. Each group contained 4 wells. To assess chemosensitivity to 5-fluoro-uracil, cells cultured for 24 h were incubated with different concentrations of 5-fluorouracil for another 48 h. Then cells were treated with MTT as described earlier. The cell survival value index was calculated as [A570(5-fluorouracil+)/A570(5-fluorouracil–)]×100%.
Western blotting Cells were lysed in RIPA buffer [1% NP-40, 0.1% sodium dodecylsulfate (SDS), 0.5% deoxycholic acid (DOC), 150 mmol/L NaCl, 10 mmol/L Tris-HCl, and phenylmethylsulfonyl fluoride(PMSF)] at 4 °C for 30 min. An equal volume of lysate was electrophoresed with SDS-polyacrylamide gel electrophoresis (PAGE; 12%). The separated proteins in the gel were transferred to the nitrocellulose membrane. The membranes were blocked with Tris-buffered saline plus 0.1% Tween-20 (TTBS) containing 5% non-fat milk for 1 h, incubated with the appropriate primary antibodies [anti Bcl-2 (Santa Cruz, USA); anti-β-actin (Sigma)] in TTBS containing 5% non-fat milk for 2 h, followed by incubation with secondary antibodies for 1 h. Finally, the proteins were visualized by enhanced chemiluminescence reagents (Pierce, USA) and the specific bands were recorded on X-ray film.
Statistical analysis All experiments were performed in triplicate. Student’s t-test was used to determine the statistical significance of the data obtained. P<0.05 was taken to represent a statistically significant difference between means.
Results
Construction of shRNA-expressing vectors with hTERT promoter We constructed an hTERT promoter-driven shRNA-expressing vector (pBlock-TRTP-shRNA-mpA). The shRNAs targeting LUC, EGFP, Bcl-2 and negative control were separately cloned into pBlock-TRTP-shRNA-mpA, and the corresponding plasmids were named pBlock-TRTP-shLUC-mpA, pBlock-TRTP-shEGFP-mpA, pBlock-TRTP-shBcl-2-mpA and pBlock-TRTP-shControl-mpA, respec-tively. All constructs were sequence-verified.
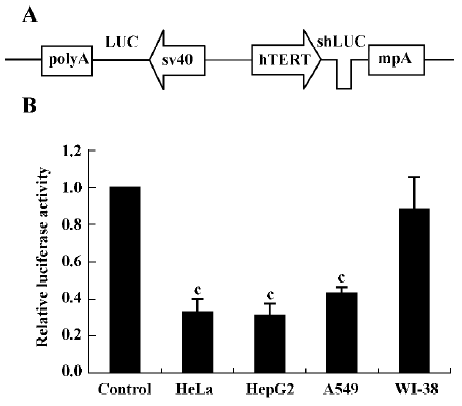
Effect of shRNA on exogenous reporter expression We first tested whether hTERT promoter-driven shLUC could inhibit the expression of firefly luciferase. To facilitate the analysis of shRNA efficacy, we constructed the pBlock-TRTP-shLUC-mpA-SV40-luciferase (pBshLUC, Figure 1A) and pBlock-TRTP-shControl-mpA-SV40-luciferase (pBLUC) vectors. Cells were transfected with the Fr-luc-expressing vectors (pBshLUC or pBLUC) and the Re-luc-expressing vector pRL-sv40. Luciferase activity was determined by a dual luciferase assay 48 h after transfection. The relative luciferase activity was defined as the ratio of Fr-luc activity to Re-luc activity. The mean pBshLUC/pBLUC ratios were 0.327±0.070, 0.308±0.063 and 0.426±0.034 in HeLa, HepG2 and A549 tumor cells, respectively (P<0.01 vs control; Figure 1B), whereas the mean pBshLUC/pBLUC ratio was 0.880±0.172 in WI-38 cells.
Next we investigated the inhibitory effects of hTERT promoter-driven shRNA targeted to another exogenous reporter, EGFP. We also constructed pBlock-TRTP-shEGFP-mpA-cmv-EGFP (pBshEGFP; Figure 2A) and pBlock-TRTP-shControl-mpA-cmv-EGFP (pBEGFP) vectors. The concentration of EGFP transcripts was determined by RT-PCR, normalized to the concentration of β-actin mRNA. We found that the concentration of EGFP transcripts decreased in HepG2, HeLa and A549 cells transfected with pBshEGFP, relative to controls, whereas there was no change in WI-38 normal cells (Figure 2B).
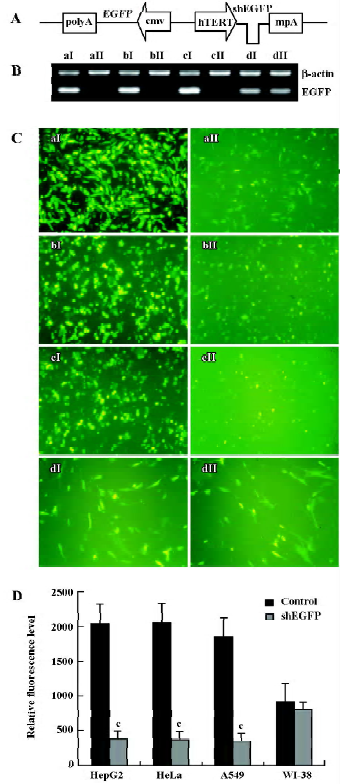
The inhibitory effects of shEGFP on EGFP protein were evaluated by fluorescence microscopy and FACS. The relative level of EGFP fluorescence did not change in WI-38 cells, but dropped markedly in HepG2, HeLa and A549 cells transfected with pBshEGFP, as determined by direct visualization under a fluorescence microscope (Figure 2C) and FACS analysis (P<0.01; Figure 2D). The results were consistent with those at the mRNA level.
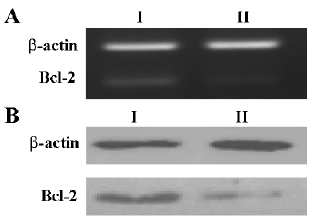
Effect of shRNA on Bcl-2 expression We next tested whether the shRNA controlled by the hTERT promoter worked on the endogenous gene Bcl-2. HeLa cells were transfected with pBlock-TRTP-shBcl-2-mpA (pBshBcl-2) or pBlock-TRTP-shControl-mpA (pBControl). For establishing HeLa cells stably expressing shRNA, cells were treated with 2 µg/mL puromycin for 14 d, and after that cells continued to be cultured with 1 µg/mL puromycin. The level of Bcl-2 expression did not change in HeLa cells stably transfected with pBControl relative to that in untransfected cells (data not shown), but was markedly reduced in cells stably transfected with pBshBcl-2, as evaluated by RT-PCR and Western blotting (Figure 3A, 3B).
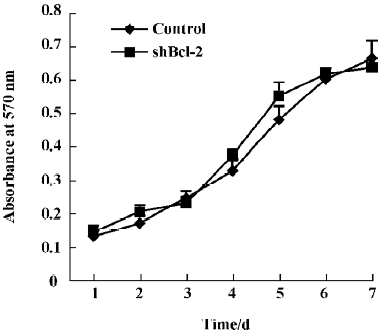
Effect of Bcl-2 expression on proliferation and cell cycle The proliferation rates of HeLa cells stably expressing shBcl-2 and shControl were similar, as determined by MTT chronometry (Figure 4). We also used FACS to analyze the cell cycle distribution profile. Inhibition of Bcl-2 did not influence the cell cycle profile (data not shown), suggesting that the Bcl-2 gene did not significantly affect cell growth or viability.
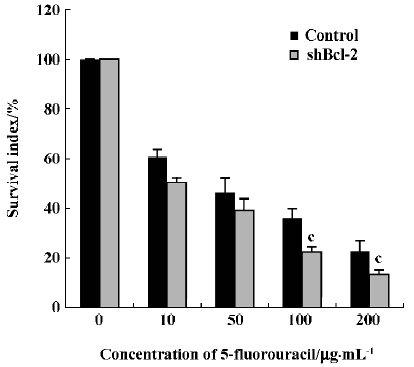
Effect of Bcl-2 expression on chemosensitivity to 5-fluorouracil Because the Bcl-2 gene has been found to play an important role in resistance to chemotherapy in many tumors, we next investigated if inhibition of Bcl-2 by RNAi affected the sensitivity of HeLa cells to the anti-tumor drug 5-fluorouracil. The survival index in HeLa cells stably expressing shBcl-2 was markedly reduced with the addition of 100 µg/mL or 200 µg/mL 5-fluorouracil (0.225±0.020 vs 0.359±0.039, 0.125±0.019 vs 0.228± 0.040; P<0.05, Figure 5) relative to controls.
Discussion
Utilization of a promoter that is predominantly active in tumor cells would be an ideal strategy for restricting therapeutic gene expression. Telomerase is a particularly attractive target because approximately 90% of tumor cells have telomerase activity, whereas most normal cells do not express that enzyme. In human cells, telomerase is under the transcriptional control of the hTERT gene, whose product is the catalytic subunit of the enzyme[16]. Transcriptional activity of the hTERT promoter is highly specific to telomerase-positive tumor cells. Therefore, the hTERT promoter has been widely used in targeting cancer gene therapy to restrict the expression of exogenous genes to various cancer cells[12–14]. A recent report also showed that the hTERT promoter might be a good, safe candidate for gene therapy[15]. To determine whether the hTERT promoter can be used to produce shRNA and silence gene expression in a tumor-specific fashion, we constructed a vector in which shRNA in the form of a small hairpin structure was placed under the control of the 403 bp hTERT promoter followed by a minimal polyA (mpA) sequence. A similar mpA has been used successfully for CMV and SP-C promoter-driven shRNA[6,11].
In the present study, we demonstrated that hTERT promoter-driven shRNA could depress the expression of the exogenous reporter genes (firefly luciferase and EGFP) in different tumor cells (HeLa, HepG2 and A549) but not normal cells (WI-38). WI-38 cells are human fetal lung fibroblasts, which have no endogenous hTERT expression[16], and are often used as representative normal cells[17,18]. hTERT promoter-driven shLUC decreased luciferase expression by approximately 70% according to the dual luciferase assay. The efficacy was similar to that of chemically synthesized siRNA or other DNA vector-based shRNA targeted to luciferase[8]. shEGFP driven by the hTERT promoter depressed the mRNA and protein expression of EGFP to the same level. This reveals that shRNA synthesized by the hTERT promoter silences gene expression in a sequence-specific manner through targeting mRNA for endonucleolitic cleavage.
We next tested whether the hTERT promoter-driven shRNA worked on the endogenous gene Bcl-2. Bcl-2 is an apoptotic inhibitor that may contribute to therapeutic resistance. It is the first oncogene found to function through production of an inhibitor of programmed cell death[19]. Overexpression of Bcl-2 is common in many human cancers, and contributes to increased resistance to chemotherapy[20–22]. Bcl-2 antisense oligonucleotides can increase the anticancer drug sensitivity of tumor cells[23,24]. Recently, successful attempts were made to silence Bcl-2 using RNAi techniques[9,25]. RNAi techniques have been repeatedly proven to be more robust than antisense techniques because they work more often, typically decrease expression of a gene to lower level, and also act at concentrations below those required for antisense techniques[25]. Therefore, antisense strategies may be replaced by RNAi approaches. Our study showed that shBcl-2 driven by the hTERT promoter could depress the expression of Bcl-2 with respect to both mRNA and protein levels. Suppression of Bcl-2 did not influence cell growth, but increased chemosensitivity to 5-fluorouracil in HeLa cells. Our results regarding the significant biological effects of tumor-specific RNAi against the Bcl-2 gene might be considered promising for future RNAi-based therapeutic approaches in HeLa cells or other malignant cell types. Using this strategy, some oncogenes could also be depressed in tumor cells without interfering with other cell systems.
In conclusion, we report here the first tumor-specific RNAi system using the hTERT promoter. We demonstrated that hTERT promoter-driven shRNA expression could effectively and specifically silence gene expression in different tumor cells but not in normal cells in vitro. We also showed that inhibition of Bcl-2 expression with this system could increase chemosensitivity to 5-fluorouracil in HeLa cells. This system might be a good candidate for future RNAi therapy.
References
- Wall NR, Shi Y. Small RNA: can RNA interference be exploited for therapy? Lancet 2003;362:1401-3.
- Sui G, Soohoo C. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA 2002;99:5515-20.
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550-3.
- Robinson R. RNAi therapeutics: how likely, how soon? PLoS Biol 2004;2:E28.
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature 2004;431:371-8.
- Gou D, Narasaraju T, Chintagari NR, Jin N, Wang P, Liu L. Gene silencing in alveolar type II cells using cell-specific promoter in vitro and in vivo. Nucleic Acids Res 2004;32:e134.
- Tiscornia G, Tergaonkar V, Galimi F, Verma IM. CRE recombinase-inducible RNA interference mediated by lentiviral vectors. Proc Natl Acad Sci USA 2004;101:7347-51.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494-8.
- Cioca D, Aoki Y, Kiyosawa K. RNA interference is a functional pathway with therapeutic potential in human myeloid leukemia cell lines. Cancer Gene Ther 2003;10:125-33.
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA 2003;100:9779-84.
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002;20:1006-10.
- Komata T, Kanzawa T, Kondo Y, Kondo S. Telomerase as a therapeutic target for malignant gliomas. Oncogene 2002;21:656-63.
- Jacob D, Davis JJ, Zhang L, Zhu H, Teraishi F, Fang B. Suppression of pancreatic tumor growth in the liver by systemic administration of the TRAIL gene driven by the hTERT promoter. Cancer Gene Ther 2005;12:109-15.
- Song JS, Kim HP, Yoon WS, Lee KW, Kim MH, Kim KT, et al. Adenovirus-mediated suicide gene therapy using the human telomerase catalytic subunit (hTERT) gene promoter induced apoptosis of ovarian cancer cell line. Biosci Biotechnol Biochem 2003;67:2344-50.
- Groot-Wassink T, Aboagye EO, Wang Y, Lemoine NR, Keith WN, Vassaux G. Noninvasive imaging of the transcriptional activities of human telomerase promoter fragments in mice. Cancer Res 2004;64:4906-11.
- Casillas MA, Brotherton SL, Andrews LG, Ruppert JM, Tollefsbol TO. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene 2003;316:57-65.
- Seo E, Abei M, Wakayama M, Fukuda K, Ugai H, Murata T, et al. Effective gene therapy of biliary tract cancers by a conditionally replicative adenovirus expressing uracil phosphoribosyltransferase: significance of timing of 5-fluorouracil administration. Cancer Res 2005;65:546-52.
- Kuo PL, Hsu YL, Lin TC, Lin CC. The antiproliferative activity of prodelphinidin B-2 3'-O-gallate from green tea leaf is through cell cycle arrest and Fas-mediated apoptotic pathway in A549 cells. Food Chem Toxicol 2005;43:315-23.
- Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev 1999;13:1899-911.
- Nicolson NL, Talpaz M, Nicolson GL. Chromatin nucleoprotein complexes containing tightly bound c-abl, p53 and bcl-2 gene sequences: correlation with progression of chronic myelogenous leukemia. Gene 1996;169:173-8.
- Sarac S, Akyol MU, Kanbur B, Poyraz A, Akyol G, Yilmaz T, et al. Bcl-2 and LMP1 expression in nasopharyngeal carcinomas. Am J Otolaryngol 2001;22:377-82.
- Khabir A, Ghorbel A, Daoud J, Frikha M, Drira MM, Laplanche A, et al. Similar BCL-X but different BCL-2 levels in the age groups of North African nasopharyngeal carcinomas. Cancer Detect Prev 2003;27:250-5.
- Lopes de Menezes DE, Hu Y, Mayer LC. Combined treatment of Bcl-2 antisense oligodeoxynucleotides (G3139), p-glycoprotein inhibitor (PSC833), and sterically stabilized liposomal doxorubicin suppresses growth of drug resistant growth of drug-resistant breast cancer in severely combined immunodeficient mice. J Exp Ther Oncol 2003;3:72-82.
- Rudin CM, Kozloff M, Hoffman PC, Edelman MJ, Karnauskas R, Tomek R, et al. Phase I study of G3139, a bcl-2 antisense oligonucleotide, combined with carboplatin and etoposide in patients with small-cell lung cancer. J Clin Oncol 2004;22:1110-7.
- Lima RT, Martins LM, Guimaraes JE, Sambade C, Vasconcelos MH. Specific downregulation of bcl-2 and xIAP by RNAi enhances the effects of chemotherapeutic agents in MCF-7 human breast cancer cells. Cancer Gene Ther 2004;11:309-16.
