Amino acid 1–209 is essential for PDX-1-mediated repression of human CMV IE promoter activity
Introduction
Pancreatic-duodenal homeobox factor-1 (PDX-1), also referred to as ipf-1[1], stf-1[2], and idx-1[3], is a homeodomain transcription factor with a molecular weight of 30 kDa, which was cloned by several laboratories. It plays a key role in regulating both pancreatic development and the differentiation of progenitor cells into the β-cell phenotype[4,5]. The expression of PDX-1 in the developing pancreas is maintained throughout development and provides both spatial and temporal contributions to the commitment of endoderm to a pancreatic phenotype[6]. Homozygous disruption of the PDX-1 gene results in pancreatic agenesis in mice and in humans[7–9]. Moreover, PDX-1 only expresses in pancreas islet β and δ cells in adults, which regulates the transcription of several genes that are essential for glucose-sensing and insulin production, including insulin[10,11], islet amyloid polypeptide[12,13], glucokinase[14], glucose transporter-2[15], and somatostatin[16]. The main DNA sequence that PDX-1 recognizes is TAAT, which is known as A-box.
PDX-1 protein contains four main domains. The first one is the transactivation domain (1–79 aa), which consists of three highly conserved sequences, subdomain A (13–22 aa), B (32–38 aa), and C (60–73 aa)[17]. These subdomains are required for the synergistic activation of insulin enhancer-mediated transcription by PDX-1 together with E2-5 and E47 proteins[10,18]. The second domain (80–149 aa) contains a pentapeptide motif FPWMK, which is very similar to the Antennapedia class of the homeodomain proteins’ motif YPWMK, and is absolutely essential for the cooperative binding of PDX-1 with Pbx to activate the somatostatin promoter in pancreatic cell lines[16,19]. The third domain is homeodomain (150–209 aa), which mediates the binding of PDX-1 with DNA and other proteins. It also mediates the nuclear localization of PDX-1, which is mainly due to an Antennapedia-like protein transduction domain (188–203 aa)[20,21]. However, the function of the last domain (210–282 aa) is still not clear.
Human cytomegalovirus (CMV) is a member of the β-herpes virus group, and is the viral agent most frequently associated with congenital infections in man. It has been reported that CMV infection is associated with many autoimmune diseases, such as lupus erythematosus[22], rheumatoid arthritis[23], and insulin-dependent diabetes mellitus (IDDM, type 1 diabetes)[24–26]. Expression of the human CMV immediately early (IE) gene is critical for productive viral replication. We used Patch search database (which searches for potential transcription factor binding sites in sequences with the pattern search program using TRANSFAC 6.0 public sites) to search potential transcription factor binding sites in CMV IE promoter and found many homeoprotein binding sites, including HOX, MEIS1, and PDX-1. Chao et al[27] have reported that PDX-1 can bind to CMV IE promoter and thus regulate its activity. They have identified a 45-bp element in CMV IE promoter, which contains multiple putative homeoprotein binding motifs, and also demonstrated the physical association between PDX-1 and the 45-bp CMV region using gel shift assays. They further determined that PDX-1 represses the CMV IE-promoter activity in HEK293 cells. However, they did not show the function that different domains of PDX-1 played in such interaction.
In this article, we investigated the roles of the domains of PDX-1 in the regulation of CMV IE promoter. Here we constructed and expressed a series of mutants lacking one or more of the four domains of wild type PDX-1 and full-length PDX-1, carrying EGFP fused to the C-terminus of the gene. Electrophoretic mobility shift assay (EMSA) results from transfected HEK293 cells showed that domain 3 is necessary and sufficient for binding to the CMV IE promoter. Luciferase assays demonstrated that full-length PDX-1 can significantly down-regulate the Human CMV IE-dependent transcription both in HEK293 and HeLa cells. We concluded that PDX-1 domain 3 was necessary for binding to CMV IE promoter, but the inhibition function required other domains to be present. The presence of PDX-1 domain 2, and 3 at the same time was essential for the negative regulation, and domain 1 could stabilize the multiprotein complex formed by PDX-1 and other proteins. So, P123 (PDX1 domain 1, 2, & 3) caused greater inhibition than that of wild type PDX-1.
Materials and methods
Cell culture and transfection HEK293, HeLa, and β-TC-6 were obtained from American Type Culture Collection; 293 and HeLa were grown in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, San Diego, CA, USA) containing 10% NCS; and β-TC-6 were grown in high glucose DMEM containing 15% FBS. Cells were grown to 50%–80% confluence in 24-well plates. Transient transfections were performed using lipofectMINE 2000 (Invitrogen, San Diego, CA, USA) according to the manufacturer’s recommen-dation, and the nuclear was stained by 4',6-Diamidino-2-phenylindole (DAPI, Sigma Chemical Co, St Louis, MO.USA).
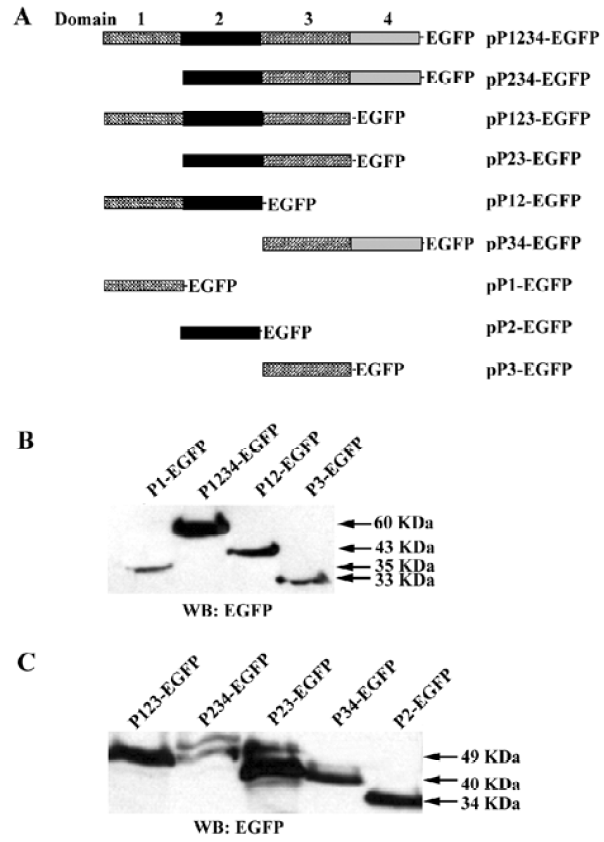
Plasmid construction The plasmid pIpf-1, which contains a murine PDX-1 gene, was a generous gift from Dr S Ferber (Institute of Endocrinology, Sheba Medical Center, Tel Hashomer, Israel). The full length PDX cDNA and PDX cDNA fragments were amplified from pIpf-1. The PCR products were digested by Mun I (designed in the 5' primers) and Kpn I (designed in the 3' primers), and were subcloned into pEGFP-N2 (Clontech) in the EcoR I and Kpn I sites. The resulting vectors were named pP1234-EGFP, pP123-EGFP, pP234-EGFP, pP12-EGFP, pP23-EGFP, pP34-EGFP, pP1-EGFP, pP2-EGFP, pP3-EGFP (numbers indicate the exact domains of PDX with EGFP fused at the C-terminus). The full-length luciferase Open Reading Frame (ORF) was digested from pGL3-Basic (Promega, Madison, WI, USA), and inserted into the multiple cloning sites of pcDNA3.0(+) using the Xho I and Xba I sites to generate pCMV-Luc.
RNA interference Oligonucleotides specifically targeted 430–448 of murine PDX-1 gene (designed by Wistar Institute siRNA selector) were cloned into the Bgl II and Hind III sites of the pSuper-vector to generate pSuper-Pdx i. The synthesized oligonucleotides containing BamH I and Hind III overhangs were as follows (the sequences shown in lowercases at 5' and 3' ends were BamH I and Hind III over-hangs, and the sequence in the middle was the loop of the RNAi oligonucleotides): forward, 5'-gatccccCCCGAGGA AAACAAGAGGAttcaagagaATCCTCTTGTTTTCCTCG-GGttttta-3'; reverse, 5'-agcttAAAAACCCGAGGAAAAC-AAGAGGATtctcttgaaTCCTCTTGTT TTCCTCGGGggg-3'.
Western blotting HEK 293 cells were transfected with the indicated expression plasmids, β-TC-6 cells were transfected with pSuper-Pdx i or pSuper-null, and cells were harvested after 24 h for western blot analysis. The samples were resolved on a 10% polyacrylamide gel, transferred to nitrocellulose, blocked with 5% (w/v) milk powder in TBST (20 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 0.05% (v/v) Tween 20), incubated with primary antisera to EGFP, mPDX-1, or mGAPDH (Santa Cruz Biotechnology) at a 1:200 dilution. After washing three times in TBST, they were incubated with horseradish peroxidase-conjugated secondary antibody at a 1:1000 dilution. Finally, the bands were detected using chemiluminescence (ECL; Amersham Biosciences).
Luciferase assay A dual luciferase assay was carried out according to manufacturer’s instructions (Promega). For the overexpression assays, 293 or HeLa cells were cotrans-fected with pCMV-Luc, the indicated expression vectors (ie, pEGFP-N2, pP1234-EGFP, pP123-EGFP, pP234-EGFP, pP12-EGFP, pP23-EGFP, pP34-EGFP, pP1-EGFP, pP2-EGFP, pP3-EGFP), and pRL-TK (Promega, to normalize for transfection).
For the RNA interference assay, β-TC-6 cells were co-transfected with pCMV-Luc, pRL-TK, and pSuper-Pdx i or not. Cells were harvested 48 h after transfection and luciferase activity were measured as relative light unit with a luminomer (Luma LB9507, EG & G Berthold, Germany). All experiments were performed in triplicate.
The means±SD of a total number of analyzed samples are indicated on the figures. The significance of the effects of various treatments as compared with untreated control was evaluated by paired Student’s t-test at the 95% confidence level.
Electrophoretic mobility shift assay A 3' biotin labeled 55-bp double-stranded CMV sequence (-598–-543, designed under the putative binding site search results and including Chao’s 45-bp CMV region) was used as the DNA probe for EMSA: 5'-GGCATTGATTATTGACTAGTTATTAATAGT-AATCAATTACGGGGTCATTAGTTCA-biotin-3' (only the forward sequence was shown). 293 cells were grown to 50%–80% confluence in 6-well plates, and transfected with 4 μg/well of pEGFP-N2, pP1234-EGFP, pP123-EGFP, pP234-EGFP, pP12-EGFP, pP23-EGFP, pP34-EGFP, pP1-EGFP, pP2-EGFP, or pP3-EGFP. Forty-eight hours later nuclear protein extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). EMSA were performed using LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s instructions.
Putative binding site search We used the program Patch (www.gene-regulation.com, formerly known as PatSearch) to analyze the CMV IE promoter. It had one putative PDX-1 homeoprotein binding site, and several putative homeoprotein family protein binding sites.
Results
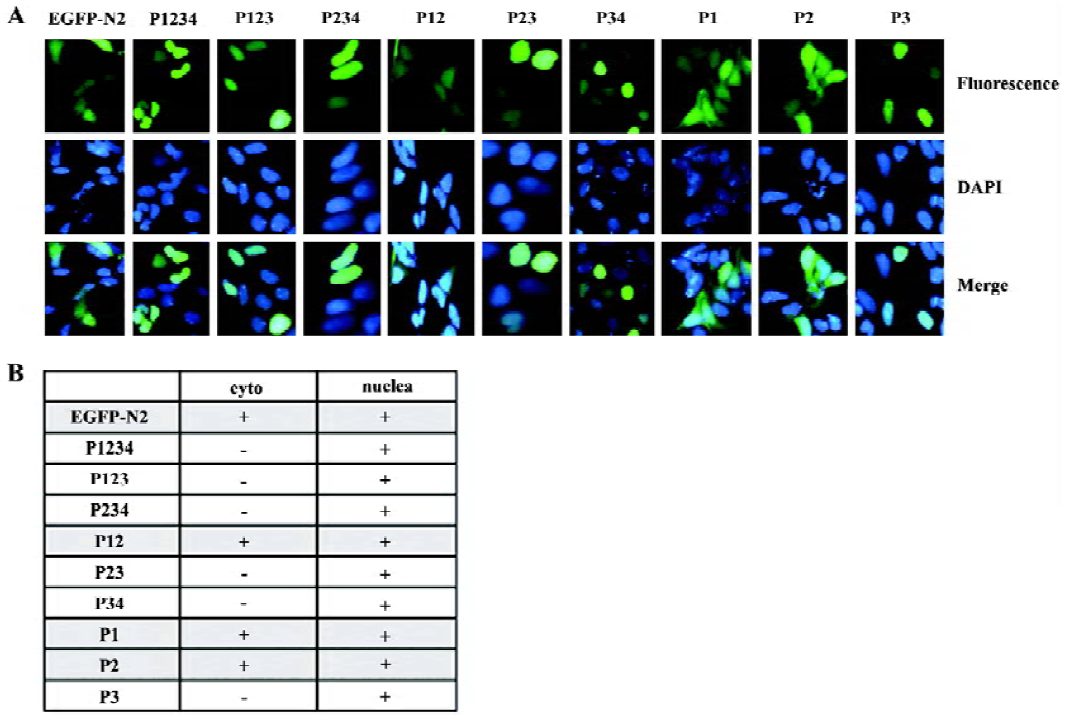
Cellular location of PDX-1 mutants First, a series of expression vectors carrying the full-length or truncated (lacking one or more domains) PDX-1, with EGFP fused at the C-terminus, were constructed. A total of nine vectors were named as pP1234-EGFP, pP234-EGFP, pP123-EGFP, pP23-EGFP, p12-EGFP, pP34-EGFP, pP1-EGFP, pP2-EGFP, pP3-EGFP (Figure 1A). HEK293 cells were transfected with different expression vectors described above and pEGFP-N2 was used as control. Cellular localization of the expressed proteins was examined using fluorescence microscopy 24 h after transfection, and the molecular weight of the series mutants were identified by Western blot (Figure 1B, 1C). Domain 3 is necessary for nuclear localization of the expressed proteins and EGFP-N2, and those mutants without domain 3 were distributed all through the cells (Figure 2).
PDX-1 downregulated human CMV IE promoter activity in both 293 and HeLa cell Using the Patch program, we found a 55-bp region (-598 to -543, which included the whole sequences of Chao’s 45-bp CMV region) in the human CMV-IE promoter carrying one putative PDX-1 homeoprotein binding sites, and several other putative binding sites of homeo-protein family members, such as HOXA and MEIS1. This suggested that PDX-1 might interact with CMV IE promoter and regulate its transcription.
To study the regulation of CMV IE promoter by PDX-1, 293 and HeLa cells were co-transfected with CMV IE luciferase reporter vector (pCMV-Luc) and PDX-1 expression vector pP1234-EGFP, respectively. Compared to the mock controls (pEGFP-N2), overexpression of PDX-1 resulted in a 41% decrease of CMV promoter activity in the 293 cells (P<0.05) and 43% decrease in the HeLa cells (P<0.05)(Figure 3A).
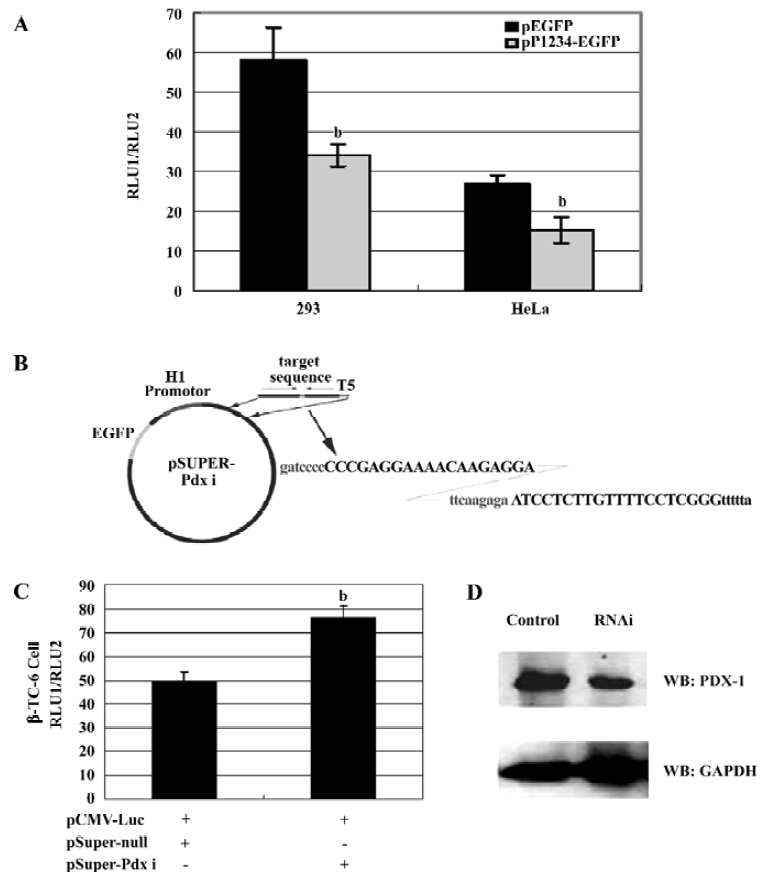
Specific knockdown of the endogenous PDX-1 expression significantly decreased its repression on CMV IE promoter Further evidence supporting the role for PDX-1 came from experiments using RNAi techniques to abolish PDX-1 expression. PDX-1 shRNA expression vectors were generated in the pSuper-background and driven by H1 promoter (Figure 3B). As shown in Figure 3C, β-TC-6 cells were cotransfected with pCMV-Luc and pSuper-Pdx i or pSuper-null constructs and achieved 40%–50% transfection efficiency (shown by EGFP reporter vector). 24 h after trans-fection, a 1.6-fold (P<0.05) increase in pCMV-Luc reporter was observed in β-TC-6 cells in the presence of pSuper-Pdx i construct, when compared to the cells transfected with pSuper-null construct. In addition, the expression of the PDX-1 protein was significantly reduced compared to cells transfected with the empty RNAi vector (50% reduction) (Figure 3D). Because only half of the cells were transfected, we estimated the efficiency of shRNA-mediated reduction of PDX-1 might be up to 70% in transfected cells. These results suggest that it is feasible to apply RNAi technology to study the repression of PDX-1 on CMV IE promoter in vitro.
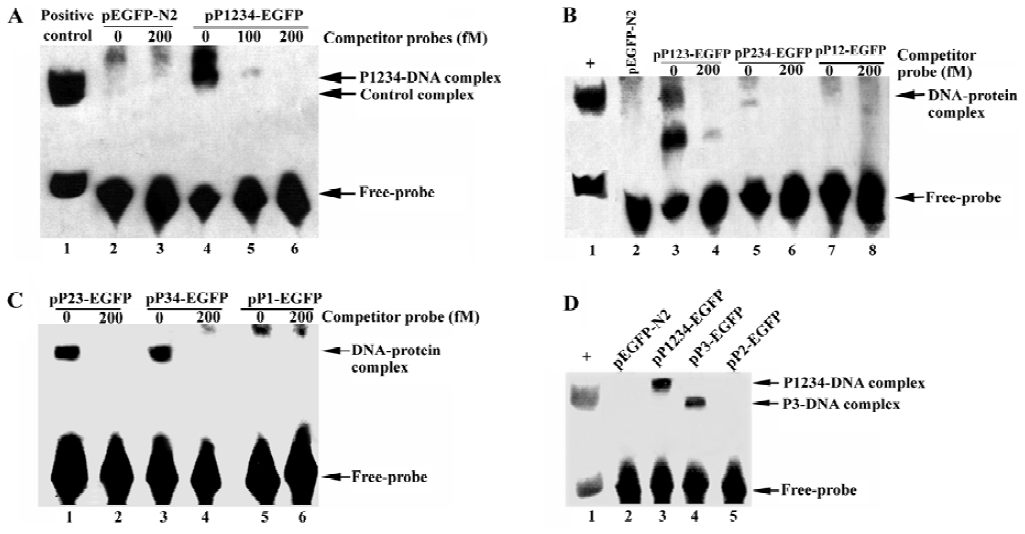
PDX-1 domain 3 necessary for binding to CMV IE promoter but inhibition function also required PDX-1 domain 1/2 be present To explore the potential role of PDX-1 in the regulation of CMV IE promoter, we applied EMSA to evaluate the binding status of wild type or truncated mutant PDX-1 to CMV IE promoter. First, we transfected HEK293 cells with the following expression vectors: pEGFP-N2, pP1234-EGFP, pP123-EGFP, pP234-EGFP, pP12-EGFP, pP23-EGFP, pP34-EGFP, pP1-EGFP, pP2-EGFP, and pP3-EGFP. Nuclear extracts of HEK 293 cells transfected with each of the expression vectors were then incubated with the 55-bp biotin-labeled CMV IE promoter probe. The results showed that there were major DNA-protein complexes when the CMV IE promoter probe incubated with P1234, P123, P234, P23, P34, or P3 nuclear extracts, all of which carried domain 3. However, no obvious DNA-protein complexes could be observed when the probe was incubated with EGFP, P12, P1, or P2 nuclear extracts, in which domain 3 was absent (Figure 4). To further confirm the binding specificity of the complex to CMV IE promoter, we performed a competition assay with unlabeled CMV IE promoter probe. The competitor oligonuleotides specifically reduced the formation of the DNA-protein complex (Figure 4A, lane 5). These results indicated that the protein complex bound specifically to the CMV IE promoter. We concluded from the observations that domain 3 of the PDX-1 protein is necessary and sufficient to confer DNA binding activity.
We further investigated the regulation of CMV IE promoter by different PDX-1 mutants using luciferase reporter assay in HEK293 cells. Each construct was cotransfected with pCMV-Luc and pRL-TK. The results showed that overexpression of P123 caused a 56.70% (P<0.05) decrease in promoter activity compared with normal CMV IE promoter activity, a 38.52% (P<0.05) decrease in P234 transfection, and a 27.28% decrease (P<0.05) in P23 transfection (Figure 5). No significant effects were observed when P12, P34, P1, P2, P3 overexpressed. According to the functions of each domain, we concluded that the absence of DNA binding domain (PDX-1 domain 3) could result in the total loss of repression activities.
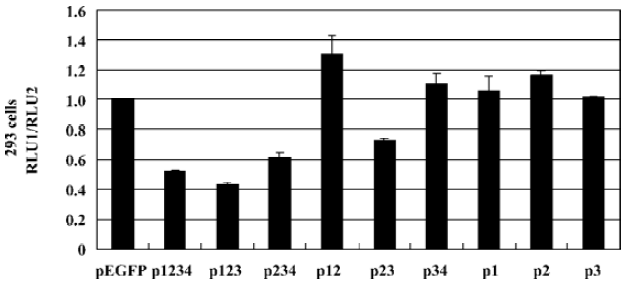
We noticed that PDX-1 domain 3 was sufficient for the binding activities of PDX-1 proteins, and P34, P3 showed specific DNA-protein complexes in EMSA. However, no promoter repression was observed in the transfection of P34 and P3, which suggested that binding of PDX-1 protein to the CMV promoter alone was not sufficient to inhibit CMV transcriptional activity. Furthermore, P23 caused a 27.28% decrease while P123 caused greater inhibition, which was close to wild type PDX-1. And we found that the DNA-protein complex formed by probe DNA and P3 nuclear extracts was much smaller than that of P1234 nuclear extracts. Based on our knowledge of the functions of the four domains, we hypothesized that there should be a multi-protein complex rather than PDX-1 alone acting as a repression unit, which inhibits the expression activity of CMV IE promoter. And the putative multi-protein complex might contain P300, Pbx, Prep1, E2-5, and E47, etc, which were known to interact with PDX-1 through domain 1 and domain 2.
Discussion
The human CMV IE promoter is one of the most commonly used promoters for overexpressing recombinant genes in a wide range of mammalian cells. Previous studies have found that a cellular homeoprotein, PDX-1, could bind to CMV IE promoter and negatively regulated its transcriptional activity in HEK 293 cell lines[27]. In the present article, we constructed eight PDX-1 truncated mutants in order to determine the molecular mechanism of PDX-1 that regulates CMV IE promoter. Through the dual-reporter and EMSA assay, we confirmed that domain 3 of PDX-1 is essential for its nuclear localization and DNA binding activity. However, PDX-1 domain 3 and domains 34 showed no transcriptional repression activities in luciferase assay. These results indicated that PDX-1 binding alone was not sufficient to repress the transcription activity of CMV IE promoter.
Further study showed that both domain 1 and domain 2 contributed to PDX-1’s repressive activity to the CMV IE promoter in the presence of domain 3. And the bands of DNA-protein complex formed by probe DNA and P3-transfected cellular nuclear extracts is much smaller than that formed by probe DNA and full-length PDX-1-transfected cellular nuclear extracts. As it has been reported that PDX-1 domain 1 and domain 2 interact with other proteins such as P300, Pbx, Prep1, E2-5, and E47[10,18,28–30], we suspected that a multi-protein complex involved in protein interaction with domain 1 and 2 was required for the negative regulation of CMV IE promoter mediated by PDX-1.
Chao et al[27] first discovered a similar phenomenon. They constructed 10 mutations of a 45-bp CMV IE promoter as we have described (Figure 6). They found that site 6 and site 10 might be PDX-1 binding sites, and the mutations of either of the two sites resulted in partial loss of PDX-1 negatively-regulate activity to CMV IE promoter. Furthermore, they found that, although site 9 showed no relationship with PDX-1 binding activity, PDX-1 lost its negatively regulating activity when it mutated.
In our work, we used the Patch program to search the CMV IE promoter sequence and found that site 10, which is in the (-) chain (Figure 6), is the only PDX-1 binding site in this sequence and the complementary sequence of site 9 in the (+) chain (Figure 6) is the cAMP response element binding protein (CREB) binding site.
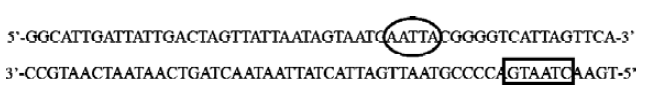
It has been reported that transcriptional activity of PDX-1 required recruitment of P300[28]. The C-terminal of P300 interacted directly with PDX-1 between amino acids 1 and 143, which is exactly PDX-1 domain 1 and 2[29], and this protein complex was supposed to be located at site 10 in the (-) chain in CMV IE promoter. It’s well known that CREB-P300/CBP complex is essential for CMV IE promoter transcription activity[31–33], which should be located at site 9, the CREB binding site, in the (+) chain in CMV IE promoter. So there might be a possibility that PDX-1, P300, CBP, and CREB could form a complex as PDX-1-P300/ CBP-CREB, binding to both (-) and (+) chain. Such a multi-protein complex could protect double-strand DNA from melting, and thus decrease transcriptional activity. However, site 9 of CMV IE promoter is the first CREB binding site. The forming of PDX-1-P300/CBP-CREB complex could also inhibit the CREB-P300/CBP complex slipping to the next CREB binding site, and it is known that the slipping was essential for the CMV IE promoter transcriptional activity. So this may explain why loss of domain 1 decreased the repression efficiency of PDX-1 upon CMV IE promoter.
According to our findings that P23 could also reduce the expression driven by CMV IE promoter, there might be another mechanism depended on PDX-1 domain 2. One of the major proteins that interacted with PDX-1 is PBX1, which has been reported to form PBX1/E2A complex interacting directly with P300/CBP complex[34]. So, our model above could also explain the repression function of domain 2.
To test our model we further constructed a CMV IE promoter mutant in which the CREB and the PDX-1 binding sites are at the same chain of DNA (+ chain). The mutant was also inserted into the multiple cloning sites of pGL3-Basic to generate pCMV+/+-Luc, and was co-transfected with pP1234-EFGP and pRL-TK. The luciferase assay results showed that PDX-1 has weak repression activity to CMV+/+ promoter (10.73% decreased), although the EMSA assay indicated the same binding activity of PDX-1 protein to CMV+/+ promoter probe (data not shown).
The 55-bp fragment used in our study is located at the CMV IE gene promoter unique region. And the importance of this unique region in human CMV IE transcription and viral replication has been investigated previously. Recombinant CMV containing deletions from positions -640 to -583, which did not contain the CREB and PDX-1 binding site, showed no significant effects on IE transcription[35]. However, a deletion of the sequence between -521 and -579, which including the preferred CREB and PDX-1 binding site, showed significant multiplicities of infection (moi)-dependent increases in recombinant CMV replication. And it had been reported that the 47-bp segment has consensus-binding sites for CREB/ATF, SP1, and YY1[36,37]. Therefore, these previous studies and our results suggested the possible involvement of PDX-1 in the CMV replication by downregulating the CMV IE promoter. And for the other cells that have no PDX-1 expression, perhaps other homeodomain proteins such as Isl, MEIS1, HOXA may bind to this region and have a similar effect to CMV IE promoter. However, more work should be carried out to explore the exact model of PDX-1 mediated downregulation of CMV IE promoter, and to discover whether the virus applied this method to shut down the expression of IE gene.
References
- Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 1993;12:4251-9.
- Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy MR. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol 1993;7:1275-83.
- Miller CP, McGehee RE, Habener JF. A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J 1994;13:1145-56.
- Stoffers DA, Thomas MK, Habener JF. Homeodomain protein IDX-1: a master regulator of pancreas development and insulin gene expression. Trends Endocrinol Metab 1997;8:145-51.
- Habener JF, Stoffers DA. A newly discovered role of transcription factors involved in pancreas development and the pathogenesis of diabetes mellitus. Proc Assoc Am Physicians 1998;110:12-21.
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 1995;121:11-8.
- Stoffers DS, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 1997;15:106-10.
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994;371:606-9.
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983-95.
- Ohneda K, Mirmira RG, Wang J, Johnson JD, German MS. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol 2000;20:900-91.
- Peers B, Leonard J, Sharma S, Teitelman G, Montminy MR. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol 1994;8:1798-806.
- Watada H, Kajimoto Y, Kaneto H, Matsuoka T, Fujitani Y, Miyazaki J, et al. Involvement of the homeodomain-containing transcription factor PDX-1 in islet amyloid polypeptide gene transcription. Biochem Biophys Res Commun 1996;229:746-51.
- Macfarlane WM, Campbell SC, Elrick LJ, Oates V, Bermano G, Lindley KJ, et al. Glucose Regulates Islet Amyloid Polypeptide Gene Transcription in a PDX1- and Calcium-dependent Manner. J Biol Chem 2000;275:15330-5.
- Watada H, Kajimoto Y, Umayahara Y, Matsuoka T, Kaneto H, Fujitani Y, et al. The human glucokinase gene beta-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes 1996;45:1478-88.
- Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol 1996;10:1327-34.
- Goudet G, Delhalle S, Biermar F, Martial JA, Peers B. Function and cooperative interactions between the homeodomain PDX-1 and Pbx, and Prp1 factors on the somatostatin promoter. J Biol Chem 1999;274:4067-73.
- Peshavaria M, Henderson E, Sharma A, Wrigh CV, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol 1997;17:3987-96.
- Glick E, Leshkowitz D, Walker MD. Transcription Factor BETA2 Acts Cooperatively with E2A and PDX1 to Activate the Insulin Gene Promoter. J Biol Chem 2000;275:2199-204.
- Andersen FG, Jensen J, Heller RS, Petersen HV, Larsson LI, Madsen OD, et al. Pax6 and Pdx1 form a functional complex on the rat somatostatin gene upstream enhancer. FEBS Letters 1999;445:315-20.
- Hessabi B, Ziegler P, Schmidt I, Hessabi C, Walther R. The nuclear localization signal (NLS) of PDX-1 is part of the homeodomain and represents a novel type of NLS. Eur J Biochem 1999;263:170-7.
- Noguchi H, Kaneto H, Weir GC, Bonner-Weir S. PDX-1 protein containing its own antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes 2003;52:1732-7.
- Sekigawa I, Nawata M, Seta N, Yamada M, Iida N, Hashimoto H. Cytomegalovirus infection in patients with systemic lupus erythematosus. Clin Exp Rheumatol 2002;20:559-64.
- Hooper M, Kallas EG, Coffin D, Campbell D, Evans TG, Looney RJ. Cytomegalovirus seropositivity is associated with the expansion of CD4+CD28- and CD8+CD28- T cells in rheumatoid arthritis. J Rheumatol 1999;26:1452-7.
- Hiemstra HS, Schloot NC, van Veelen PA, Willemen SJ, Franken KL, van Rood JJ, et al. Cytomegalovirus in autoimmunity: T cell crossreactivity to viral antigen and autoantigen glutamic acid decarboxylase. Proc Natl Acad Sci USA 2001;98:3988-91.
- Jun HS, Yoon JW. A new look at viruses in type 1 diabetes. ILAR J 2004;45:349-74.
- Pak CY, Eun HM, McArthur RG, Yoon JW. Association of cytomegalovirus infection with autoimmune type 1 diabetes. Lancet 1988;2:1-4.
- Chao SH, Harada JN, Hyndman F, Gao XQ, Nelson CG, Chanda SK, et al. PDX1, a Cellular Homeoprotein, Binds to and Regulates the Activity of Human Cytomegalovirus Immediate Early Promoter. J Biol Chem 2004;279:16111-20.
- Stanojevic V, Habener JF, Thomas MK. Pancreas duodenum homeobox-1 transcriptional activation requires interactions with p300. Endocrinology 2004;145:2918-28.
- Qiu Y, Guo M, Huang SM, Stein R. Insulin Gene Transcription Is Mediated by Interactions between the p300 Coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002;22:412-20.
- Dutta S, Gannon M, Peers B, Wright C, Bonner-Weir S, Montminy M. PDX:PBX complexes are required for normal proliferation of pancreatic cells during development. Proc Natl Acad Sci USA 2001;98:1065-70.
- Sanchez TA, Habib I, Leland Booth J, Evetts SM, Metcalf JP. Zinc finger and carboxyl regions of adenovirus E1A 13S CR3 are important for transactivation of the cytomegalovirus major immediate early promoter by adenovirus. Am J Respir Cell Mol Biol 2000;23:670-7.
- Olive DM, al-Mulla W, Simsek M, Zarban S, al-Nakib W. The human cytomegalovirus immediate early enhancer-promoter is responsive to activation by the adenovirus-5 13S E1A gene. Arch Virol 1990;112:67-80.
- Stamminger T, Fleckenstein B. Immediate-early transcription regulation of human cytomegalovirus. Curr Top Microbiol Immunol 1990;154:3-19.
- Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, et al. E2A-PBX1 Interacts Directly with the KIX Domain of CBP/p300 in the Induction of Proliferation in Primary Hematopoietic Cells. J Biol Chem 2004;279:55362-71.
- Meier JL, Keller MJ, McCoy JJ. Requirement of Multiple cis-Acting Elements in the Human Cytomegalovirus Major Immediate-Early Distal Enhancer for Viral Gene Expression and Replication. J Virol 2002;76:313-26.
- Meier JL, Stinski MF. Regulation of cytomegalovirus immediate early genes. Intervirol 1996;39:331-42.
- Sculley TB, Spelsberg TC, Pearson GR. Characterization of Epstein-Barr virus nuclear antigen(s) in different cell lines by radioimmunoelectrophoresis. Intervirol 1984;22:191-200.
